Back to Journals » Drug Design, Development and Therapy » Volume 12
Symmetric dimeric adamantanes for exploring the structure of two viroporins: influenza virus M2 and hepatitis C virus p7
Authors Mandour YM , Breitinger U , Ma C, Wang J, Boeckler FM , Breitinger HG , Zlotos DP
Received 4 December 2017
Accepted for publication 3 March 2018
Published 30 April 2018 Volume 2018:12 Pages 1019—1031
DOI https://doi.org/10.2147/DDDT.S157104
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Qiongyu Guo
Yasmine M Mandour,1 Ulrike Breitinger,2 Chunlong Ma,3 Jun Wang,3 Frank M Boeckler,4 Hans-Georg Breitinger,2 Darius P Zlotos1
1Department of Pharmaceutical Chemistry, German University in Cairo, 2Department of Biochemistry, The German University in Cairo, Cairo, Egypt; 3Department of Pharmacology and Toxicology, University of Arizona, Tucson, AZ, USA; 4Department of Pharmaceutical and Medicinal Chemistry, Eberhard Karls University of Tübingen, Tübingen, Germany
Background: Adamantane-based compounds have been identified to interfere with the ion-channel activity of viroporins and thereby inhibit viral infection. To better understand the difference in the inhibition mechanism of viroporins, we synthesized symmetric dimeric adamantane analogs of various alkyl-spacer lengths.
Methods: Symmetric dimeric adamantane derivatives were synthesized where two amantadine or rimantadine molecules were linked by various alkyl-spacers. The inhibitory activity of the compounds was studied on two viroporins: the influenza virus M2 protein, expressed in Xenopus oocytes, using the two-electrode voltage-clamp technique, and the hepatitis C virus (HCV) p7 channels for five different genotypes (1a, 1b, 2a, 3a, and 4a) expressed in HEK293 cells using whole-cell patch-clamp recording techniques.
Results: Upon testing on M2 protein, dimeric compounds showed significantly lower inhibitory activity relative to the monomeric amantadine. The lack of channel blockage of the dimeric amantadine and rimantadine analogs against M2 wild type and M2-S31N mutant was consistent with previously proposed drug-binding mechanisms and further confirmed that the pore-binding model is the pharmacologically relevant drug-binding model. On the other hand, these dimers showed similar potency to their respective monomeric analogs when tested on p7 protein in HCV genotypes 1a, 1b, and 4a while being 700-fold and 150-fold more potent than amantadine in genotypes 2a and 3a, respectively. An amino group appears to be important for inhibiting the ion-channel activity of p7 protein in genotype 2a, while its importance was minimal in all other genotypes.
Conclusion: Symmetric dimeric adamantanes can be considered a prospective class of p7 inhibitors that are able to address the differences in adamantane sensitivity among the various genotypes of HCV.
Keywords: adamantane, dimers, viroporins, p7, M2, HCV
Introduction
Viroporins are small virus-encoded proteins that are able to permeabilize membranes for ions and small molecules.1,2 This family of proteins includes M2,3 Vpu,4 the E-protein channels of coronaviruses,5 and the p7 ion channel of hepatitis C virus (HCV).6 Viroporins commonly are small transmembrane (TM) proteins that contain several identical self-oligomerizing subunits forming functional ion channels. However, sequence similarity among the viroporin family is low, and the functional channels differ in subunit size, stoichiometry, and topology.7 Viroporins are crucial for viral pathogenicity, owing to their involvement in several steps of the viral life cycle, but their main activities are in cell entry, virion assembly, and release from infected cells.8 M2 protein of influenza virus equilibrates the pH across the viral membrane during cell entry, facilitating RNA release,9,10 and across the trans-Golgi membrane of infected cells during viral maturation.11 The p7 protein of HCV is believed to act as an intracellular pH shunt, contributing to the deacidification of endosomes and virion-loaded particles and assisting in assembly and release of virions,12 as well as promoting inflammatory signaling.13
Owing to their role in the viral life cycle and their structural differences from human channels, these viral proteins are considered ideal antiviral drug targets.8 Several compounds that block viroporin ion-channel activity have been identified. Two adamantane based compounds, amantadine and rimantadine, were the first US Food and Drug Administration-approved drugs for treatment of influenza infections.14 These compounds inhibit the M2 protein by blocking its proton conductance.15,16 It has also been found that these adamantane-based compounds can block HCV p7 ion-channel activity17–19 and abolish p7-mediated inflammatory signaling in liver macrophages.13
Structure determination of viroporins under physiological conditions has been difficult, due to the hydrophobic and dynamic nature of these proteins. The few available viroporin structures revealed unexpected architecture that helped in understanding the operation of these channels.8 In addition, the binding mode of several ion-channel blockers was revealed, which provided guidance for designing other inhibitors.8 Several structures of M2 protein in complex with adamantanes have been solved. These structures showed M2 as a tetramer and suggested two possible binding models for adamantanes: the pore-binding model and the exterior interface-binding model.20,21 In the pore-binding model, one adamantane molecule physically blocks the ion channel, breaking the continuous water wires in the channel that are critical for proton conductance.21 For the exterior interface-binding model, four adamantane molecules bind allosterically at an external, lipid-facing pocket between adjacent monomers, thus inducing conformational changes in the TM helices that favor the closed state of the channel.20 It was later confirmed that the pore-binding model is the pharmacologically relevant model that accounts for the antiviral activity of amantadine and rimantadine.22,23 Unfortunately, naturally occurring drug-resistant mutations have gradually decreased these adamantanes’ inhibition ability. S31N is the predominant amantadine-resistant M2 form that represents a huge challenge to drug discovery.
The structure of p7 protein has been studied using nuclear magnetic resonance (NMR) methods24 and molecular modeling,25 showing different potential topologies and conformations. P7 is a 63-residue protein with two TM domains (TMDs) that are separated by a short hydrophilic cytosolic loop.24,25 The p7 protomer is able to oligomerize to hexameric21,24 and heptameric complexes,17 forming an ion channel. Structural studies have revealed two proposed structures for the oligomeric form of p7. The first structure proposes a funnel-like architecture, where the individual p7 monomer, i, interacts with i + 2 and i + 3 monomers, forming a hexameric assembly.26 The second structure suggests a flower-shaped protein architecture, where each unit interacts only with neighboring monomers.27 However, in both structures six adamantane molecules are proposed to bind to six equivalent hydrophobic pockets formed by residues from two adjacent monomers.
Previous studies have shown dimeric camphor derivatives, which have a cage-like structure similar to adamantane, to possess high antiviral activity against influenza. It was postulated that two camphor moieties bind to two adjacent sites of neighboring chains of M2.28 However, these compounds were not tested on M2 protein, so the exact mechanism of inhibition of these compounds is unknown. It is noted that cage-shape molecules might inhibit influenza viruses through M2-independent mechanisms.22 To further elucidate the M2 drug-binding mechanism and rule out the possibility of the exterior interface-binding model, we hereby report the design and characterization of dimeric amantadines and rimantadines. If the exterior interface-binding model is correct, the dimeric amantadines and rimantadines should have comparable or better channel-blockage activity than the monomeric amantadine and rimantadine. If the pore-binding model is correct, the dimeric amantadines and rimantadines should not be able to fit in the channel and have no channel inhibition. In contrast, the structure of p7 protein suggests the presence of several identical adamantane-binding pockets where six adamantane molecules are required to inhibit one p7 protein molecule. Accordingly, combination of two adamantane moieties in one molecule might lead to supra-additive increase in potency compared to the corresponding monovalent ligands. To test this hypothesis, we synthesized symmetric dimeric adamantane analogs of various alkyl-spacer lengths to find the optimum distance that allows simultaneous binding of the two adamantane rings. Here, we describe the design and synthesis of symmetric dimeric adamantane derivatives of various alkyl-spacer lengths and the determination of their inhibitory activity on two viroporins: M2 channels of influenza virus and the p7 protein of HCV.
Methods
General chemistry
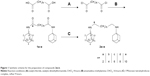
All applied starting materials were commercially available from Alfa Aesar and Sigma-Aldrich and used as received. Column chromatography: silica gel 60 (0.063–0.200 mm; Merck). Reaction and column-chromatography purification progress was monitored by TLC silica gel (60F154) aluminum sheets. Compound spots were visualized in an iodine chamber. Melting point (MP): capillary MP apparatus (Sanyo Gallenkamp, UK), uncorrected. 1H and 13C NMR spectra were recorded on a Bruker AV-400 spectrometer (1H, 400.13 MHz; 13C, 100.61 MHz). Proton chemical shifts are referenced to CHCl3 (7.26 ppm) or methanol-d4 (3.31 ppm). Coupling constants (J) are given in hertz. Carbon chemical shifts are referenced to CDCl3 (77 ppm) or methanol-d4 (49 ppm) (Figures S1–S12). Electrospray ionization (ESI) mass spectrometry (MS) was determined on an Agilent 1100 liquid-chromatography MS system in a positive mode. Elemental analyses were performed by the microanalytical section of the Institute of Inorganic Chemistry, Würzburg University, and the results were within ±0.4% of the theoretical values.
Procedure A for synthesis of compounds 1a–e
To a stirred solution of the respective dicarboxylic acid (1.4 mmol) in dry chloroform (20 mL), oxalyl chloride (0.48 mL, 5.6 mmol) was added dropwise, followed by addition of two drops of dry dimethylformamide. The reaction mixture was left to stir at room temperature (RT) for 4 hours. Toluene (20 mL) was added to the reaction mixture and evaporated under reduced pressure. The resultant acid chloride was dissolved in dry CHCl3 (3 mL) and the solution added dropwise to a stirred solution of amantadine (0.6 g, 4.0 mmol) and dry triethylamine (0.5 mL, 3.305 mmol) in CHCl3 (30 mL). The reaction mixture was left to stir for 16 hours at RT under inert conditions and washed with diluted HCl (2×30 mL). The combined organic layers were washed with water, dried over sodium sulfate, and concentrated under reduced pressure. The resulting crude product was pure enough to be used directly for the next step without further purification, as indicated by 1H NMR spectra.
N,N′-bis-(adamantan-1-yl) succinic acid diamide (1a)
Compound 1a (400 mg, 74%) was obtained as a white solid; MP: 210°C. 1H NMR (CD3OD, 400 MHz) δ (ppm) 1.61–1.67 (m, 6H, 4,6,10-adamantane H), 1.96 (d, J=2.4 Hz, 6H, 2,9,8-adamantane H), 1.99 (br s, 3H, 3,5,7-adamantane H), 2.52 (br s, 2H, H-b). 13C NMR (CD3OD, 100 MHz) δ (ppm) 30.84 (3,5,7-adamantane C), 32.05 (C-b), 37.29 (4,6,10-adamantane C), 41.90 (2,8,9-adamantane C), 51.78 (1-adamantane C), 175.85 (C-a). EI-MS: m/z 384.25 [M]+.
N,N′-bis-(adamantan-1-yl) adipic acid diamide (1b)
Compound 1b (420 mg, 74%) was obtained as white solid; MP: 268°C. 1H NMR (CDCl3, 400 MHz) δ (ppm) 1.53–1.58 (m, 2H, H-c), 1.60 (m, 6H, 4,6,10-adamantane H), 1.93 (d, J=2.5 Hz, 6H, 2,9,8-adamantane H), 2.00 (br s, 3H, 3,5,7-adamantane H), 2.03 (t, J=6.4 Hz, 2H, H-b), 5.27 (s, 1H, NH). 13C NMR (CDCl3, 100 MHz) δ (ppm) 25.09 (C-c) 29.45 (3,5,7-adamantane C), 37.27 (C-b), 36.38 (4,6,10-adamantane C), 41.66 (2,8,9-adamantane C), 51.87 (1-adamantane C), 172.11 (C-a). EI-MS: m/z 412.25 [M]+.
N,N′-bis-(adamantan-1-yl) suberic acid diamide (1c)
Compound 1c (330 mg, 54%) was obtained as a white solid; MP: 228°C. 1H NMR (CD3OD, 400 MHz) δ (ppm) 1.33–1.40 (m, 2H, d H), 1.55–1.63 (m, 2H, H-c), 1.73 (m, 6H, 4,6,10-adamantane H), 2.04 (br s, 6H, 2,9,8-adamantane H), 2.06 (br s, 3H, 3,5,7-adamantane H), 2.11 (t, J=7.5 Hz, 2H, H-b). 13C NMR (CD3OD, 100 MHz) δ (ppm) 25.65 (C-d), 28.49 (C-c), 29.52 (3,5,7-adamantane C), 36.12 (4,6,10-adamantane C), 36.48 (C-b), 40.97 (2,8,9-adamantane C), 51.87 (1-adamantane C), 172.3 (C-a). ESI-MS: m/z 441.30 [M+H]+.
N,N′-bis-(adamantan-1-yl) sebacic acid diamide (1d)
Compound 1d (430 mg, 66%) was obtained as a white solid; MP: 228°C. 1H NMR (CDCl3, 400 MHz) δ (ppm) 1.22 (br s, 4H, H-d,e), 1.47–1.55 (m, 2H, H-c), 1.60 (m, 6H, 4,6,10-adamantane H), 1.92 (d, J=2.7 Hz, 6H, 2,9,8-adamantane H), 2.03 (t, J=7.4 Hz, 5H, 3,5,7-adamantane H, H-b), 5.04 (s, 1H, NH). 13C NMR (CDCl3, 100 MHz) δ (ppm) 25.73 (C-e), 29.09 (C-d), 29.14 (C-c), 29.46 (3,5,7-adamantane C), 36.39 (4,6,10-adamantane C), 37.77 (C-b), 41.73 (2,8,9-adamantane C), 51.78 (1-adamantane C), 172.33 (C-a). EI-MS: m/z 468.30 [M]+.
N,N′-bis-(adamantan-1-yl) dodecanedioic acid diamide (1e)
Compound 1e (560 mg, 82%) was obtained as a white solid; MP: 179°C. 1H NMR (CD3OD, 400 MHz) δ (ppm) 1.34 (br s, 6H, d-f H), 1.56–1.64 (m, 2H, H-c), 1.74 (m, 6H, 4,6,10-adamantane H), 2.05 (d, J=2.5 Hz, 6H, 2,9,8-adamantane H), 2.08 (s, 3H, 3,5,7-adamantane H), 2.17 (t, J=6.4 Hz, 2H, H-b). 13C NMR (CD3OD, 100 MHz) δ (ppm) 27.28 (C-f), 30.13 (C-e), 30.36 (d C), 30.48 (C-c), 30.91 (3,5,7-adamantane C), 37.46 (4,6,10-adamantane C), 37.62 (C-b), 42.25 (2,8,9-adamantane C), 51.78 (1-adamantane C), 172.33 (C-a). ESI-MS: m/z 497.4 [M+H]+.
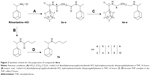
Procedure B for synthesis of compounds 3a–e
Dicarboxylic acid (0.7 mmol), 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDCI) HCl (0.423 g, 2.1 mmol), hydroxybenzotriazole (0.283 g, 2.1 mmol), and diisopropylethylamine (0.97 mL, 5.6 mmol) were dissolved in sufficient amount of dry tetrahydrofuran (THF; 20 mL). After stirring for 30 minutes, a solution of rimantadine hydrochloride (0.604 g, 2.8 mmol) in dry THF (10 mL) was added dropwise and the reaction mixture stirred overnight at RT under inert conditions. A saturated solution of NaHCO3 (100 mL) was added and the product extracted with ethyl acetate (2×40 mL) The combined organic layers were washed with water and concentrated under reduced pressure to remove the solvent. The residue was dissolved in CHCl3 (30 mL) and the solution extracted with concentrated HCl solution (2×50 mL). The chloroform phase was washed with water and dried over sodium sulfate. The solvent was evaporated under reduced pressure and the residue purified by column chromatography (dichloromethane:MeOH, 10:0.5).
N,N′-bis-(adamantan-1-yl-ethyl)-succinic acid diamide (3a)
Compound 3a (200 mg, 97%) was obtained as a white solid; MP: 191°C. 1H NMR (CDCl3, 400 MHz) δ (ppm) 0.94 (d, J=6.9 Hz, 3H, H-12), 1.43 (dd, J=26.3, 12.6 Hz, 6H, 4,6,10-adamantane H), 1.6 (m, 6H, 2,8,9-adamantane H), 1.91 (br s, 3H, 3,5,7-adamantane H), 2.42–2.52 (br s, 2H, H-b), 3.61 (m, 1H, H-11), 5.86 (s, 1H, NH). 13C NMR (CDCl3, 100 MHz) δ (ppm) 14.52 (C-12), 28.32 (3,5,7-adamantane C), 32.38 (C-b), 35.71 (1-adamantane C), 37.04 (4,6,10-adamantane C), 38.34 (2,8,9-adamantane C), 53.15 (C-11), 171.69 (C-a). EI-MS: m/z 441.20 [M+H]+.
N,N′-bis-(1-adamantan-1-yl-ethyl) adipic acid diamide (3b)
Compound 3b (510 mg, 69%) was obtained as a white solid; MP: 205°C. 1H NMR (CDCl3, 400 MHz) δ (ppm) 0.94 (d, J=6.9 Hz, 3H, H-12), 1.44 (dd, J=26.3, 12.6 Hz, 6H, 4,6,10-adamantane H), 1.67–1.51 (m, 8H, 2,8,9-adamantane H, H-c), 1.91 (br s, 3H, 3,5,7-adamantane H), 2.15 (t, J=4.6, 2H, H-b), 3.65 (m, 1H, H-11), 5.39 (s, 1H, NH). 13C NMR (CDCl3, 100 MHz) δ (ppm) 14.60 (C-12), 25.23 (C-c), 28.32 (3,5,7-adamantane C), 35.71 (1-adamantane C), 36.54 (C-b), 37.06 (4,6,10-adamantane C), 38.43 (2,8,9-adamantane C), 52.87 (C-11), 172.07 (C-a). EI-MS: m/z 468.30 [M]+.
N,N′-bis-(1-adamantan-1-yl-ethyl) suberic acid diamide (3c)
Compound 3c (200 mg, 87%) was obtained as a white solid; MP: 191°C. 1H NMR (CDCl3, 400 MHz) δ (ppm) 0.94 (d, J=6.9 Hz, 3H, H-12), 1.27–1.33 (m, 2H, H-d), 1.44 (dd, J=26.3, 12.6 Hz, 6H, 4,6,10-adamantane H), 1.52–1.67 (m, 8H, 2,8.9-adamantane H, H-c), 1.92 (br s, 3H, 3,5,7-adamantane H), 2.10 (t, J=7.5, 2H, H-b), 3.65 (m, 1H, CH-11), 5.25 (br s, 1H, NH). 13C NMR (CDCl3, 100 MHz) δ (ppm) 14.65 (12-C), 25.65 (d C), 28.32 (3,5,7-adamantane C), 28.78 (C-c), 35.71 (1-adamantane C), 36.99 (b-C), 37.07 (4,6,10-adamantane C), 38.43 (2,8,9-adamantane C), 52.74 (C-11), 172.31 (C-a). EI-MS: m/z 497.35 [M]+.
N,N′-bis-(1-adamantan-1-yl-ethyl) sebacic acid diamide (3d)
Compound 3d (200 mg, 82%) was obtained as a yellow oil. 1H NMR (CDCl3, 400 MHz) δ (ppm) 0.94 (d, J=6.9 Hz, 3H, H-12), 1.20–1.30 (m, 4H, H-d,e), 1.44 (dd, J=27.6, 12.7 Hz, 6H, 4,6,10-adamantane H), 1.51–1.68 (m, 8H, 2,8,9-adamantane H, H-c), 1.92 (br s, 3H, 3,5,7-adamantane H), 2.10 (t, J=7.5, 2H, H-b), 3.65 (m, 1H, H-11), 5.20 (br s, 1H, NH). 13C NMR (CDCl3, 100 MHz) δ (ppm) 14.65 (C-12), 25.88 (C-e), 28.32 (3,5,7-adamantane C), 29.10 (C-d), 29.20 (C-c), 35.71 (1-adamantane C), 37.07 (4,6,10-adamantane C), 37.19 (C-b), 38.43 (2,8,9-adamantane C), 52.72 (C-11), 172.45 (C-a). EI-MS: m/z 525.30 [M+H]+.
N,N′-bis-(1-adamantan-1-yl-ethyl) dodecanedioic acid diamide (3e)
Compound 3e (200 mg, 78%) was obtained as a yellow oil. 1H NMR (CDCl3, 400 MHz) δ (ppm) 0.94 (d, J=6.9 Hz, 3H, H-12), 1.19–1.28 (m, 6 H, H-d–f), 1.44 (dd, J=27.4, 12.6 Hz, 6H, 4,6,10-adamantane H), 1.67–1.51 (m, 8H, 2,8,9-adamantane H, H-c), 1.92 (br s, 3H, 3,5,7-adamantane H), 2.10 (t, J=7.5, 2H, H-b), 3.65 (m, 1H, H-11), 5.22 (br s, 1H, NH). 13C NMR (CDCl3, 100 MHz) δ (ppm) 14.65 (C-12), 25.95 (C-f), 28.43 (3,5,7-adamantane C), 29.27 (C-e), 29.38 (C-d), 29.69 (C-c), 35.71 (1-adamantane C), 37.07 (4,6,10-adamantane C), 37.24 (C-b), 38.43 (2,8,9-adamantane C), 52.72 (C-11), 172.41 (C-a). EI-MS: m/z 553.45 [M+H]+.
Procedure C for synthesis of compounds 2a–e, 4a–e, 5a
Borane THF solution (1 M, 5 mL) was added dropwise to a stirred mixture of the indicated amount of the corresponding amide in dry THF (20 mL) and heated under reflux for 9 hours under inert conditions. 2 M HCl (2 mL) was carefully added under ice cooling and the reaction mixture heated under reflux for 30 minutes. The mixture was left to cool at RT and basified with 25% aqueous ammonia under ice cooling. The product was extracted with dichloromethane (2×20 mL), and combined organic layers washed with water and dried over sodium sulfate. The solvent was evaporated under reduced pressure and the residue purified by column chromatography (chloroform:MeOH:ammonia, 100:5:1).
N,N′-bis-(adamantan-1-yl)-butane-1,4-diamine (2a)
Compound 2a (100 mg, 77%) was obtained as a white solid from 128.34 mg of 1a; MP: 52°C. 1H NMR (CD3OD, 400 MHz) δ (ppm) 1.43–1.38 (m, 2H, H-b), 1.67–1.53 (complex m, 12H, 2,4,6,8,9,10-adamantane H), 1.97 (br s, 3H, 3,5,7-adamantane H), 2.49 (t, J=7.02 Hz, 2H, H-a). 13C NMR (CD3OD, 100 MHz) δ (ppm) 29.06 (C-b), 30.98 (3,5,7-adamantane C), 37.69 (4,6,10-adamantane C), 41.0 (C-a), 42.72 (2,8,9-adamantane C), 52.21 (1-adamantane C). EI-MS: m/z 357.4 [M+H]+.
N,N′-bis-adamantan-1-yl-hexane-1,4-diamine (2b)
Compound 2b (75 mg, 54%) was obtained as a white solid from 138.43 mg of 1b; MP: 53°C. 1H NMR (CDCl3, 400 MHz) δ (ppm) 1.3–1.22 (m, 2H, H-c), 1.43–1.35 (m, 2H, H-b), 1.63–1.50 (complex m, 12H, 2,4,6,8,9,10-adamantane H), 1.99 (br s, 3H, 3,5,7-adamantane H), 2.49 (t, J=7.31 Hz, 2H, H-a). 13C NMR (CDCl3, 100 MHz) δ (ppm) 27.48 (C-c), 31.05 (C-b), 29.6 (3,5,7-adamantane C), 36.77 (4,6,10-adamantane C), 40.34 (C-a), 42.72 (2,8,9-adamantane C), 52.21 (1-adamantane C). EI-MS: m/z 385.5 [M+H]+.
N,N′-bis-adamantan-1-yl-octane-1,4-diamine (2c)
Compound 2c (113 mg, 75%) was obtained as a white solid from 140.56 mg of 1c; MP: 55°C. 1H NMR (CD3OD, 400 MHz) δ (ppm) 1.39 (br s, 4H, H-c,d), 1.48–1.59 (m, 2H, H-b), 1.68–1.77 (complex m, 12H, 2,4,6,8,9,10-adamantane H), 2.12 (br s, 3H, 3,5,7-adamantane H), 2.67 (t, J=7.6 Hz, 2H, H-a). 13C NMR (CD3OD, 100 MHz) δ (ppm) 28.32 (C-d), 30.45 (C-c), 30.47 (C-b), 30.89 (3,5,7-adamantane C), 36.77 (4,6,10-adamantane C), 40.34 (C-a), 42.72 (2,8,9-adamantane C), 52.21 (1-adamantane C). EI-MS: m/z 413.7 [M+H]+.
N,N′-bis-adamantan-1-yl-decane-1,4-diamine (2d)
Compound 2d (47 mg, 50%) was obtained as a white solid from 159.63 mg of 1d; MP: 59°C. 1H NMR (CD3OD, 400 MHz) δ (ppm) 1.39 (broad s, 6H, H-c-e), 1.49–1.57 (m, 2H, H-b), 1.67–1.80 (complex m, 12H, 2,4,6,8,9,10-adamantane H), 2.12 (br s, 3H, 3,5,7-adamantane H), 2.68 (t, J=7.7 Hz, 2H, H-a). 13C NMR (CD3OD, 100 MHz) δ (ppm) 27.43 (C-e) 29.36 (C-d), 30.76 (C-c), 29.36 (C-b), 29.57 (3,5,7-adamantane C), 36.71 (4,6,10-adamantane C), 40.40 (C-a), 42.42 (2,8,9-adamantane C), 52.21 (1-adamantane C). EI-MS: m/z 441.88 [M+H]+.
N,N′-bis-adamantan-1-yl-dodecane-1,4-diamine (2e)
Compound 2e (92 mg, 54%) was obtained as a white solid from 178.83 mg of 1e; MP: 114°C. 1H NMR (CD3OD, 400 MHz) δ (ppm) 1.22 (broad s, 8H, H-c–f), 1.33–1.41 (m, 2H, H-b), 1.52–1.67 (complex m, 12H, 2,4,6,8,9,10-adamantane H), 1.97 (br s, 3H, 3,5,7-adamantane H), 2.47 (t, J=7.6 Hz, 2H, H-a). 13C NMR (CD3OD, 100 MHz) δ (ppm) 28.6 (C-f) 30.7 (C-c–e), 31.0 (3,5,7-adamantane C), 31.21 (C-b), 37.73 (4,6,10-adamantane C), 41.182 (C-a), 42.75 (2,8,9-adamantane C), 52.21 (1-adamantane C). ESI-MS: m/z 469.40 [M+H]+.
N,N′-bis-(1-adamantan-1-yl-ethyl)-butane-1,4-diamine (4a)
Compound 4a (100 mg, 53%) was obtained as a colorless oil from 158.53 mg of 3a. 1H NMR (CDCl3, 400 MHz) δ (ppm) 0.87 (d, J=6.5 Hz, 3H, H-12), 1.38–1.42 (m, 2H, H-b), 1.42–1.55 (complex m, 6H, 4,6,10-adamantane H), 1.6 (dd, J=27.3, 11.8 Hz, 6H, 2,8,9-adamantane H), 1.90 (br s, 3H, 3,5,7-adamantane H), 1.98 (q, J=6.4, 1H, H-11), 2.36 (dt, J=11.1, 6.3 Hz, 1H, H-a), 2.67 (dt, J=11.1, 6.3 Hz, 1H, H-a). 13C NMR (CDCl3, 100 MHz) δ (ppm) 13.59 (C-12), 28.2 (C-b), 28.61 (3,5,7-adamantane C), 36.14 (1-adamantane C), 37.39 (4,6,10-adamantane C), 38.74 (2,8,9-adamantane C), 48.97 (C-a), 62.63 (C-11). ESI-MS: m/z 413.3 [M+H]+.
N,N′-bis-(1-adamantan-1-yl-ethyl)-hexane-1,6-diamine (4b)
Compound 4b (70 mg, 48%) was obtained as a colorless oil from 169.83 mg of 3b. 1H NMR (CDCl3, 400 MHz) δ (ppm) 0.87 (d, J=6.5 Hz, 3H, H-12) 1.23–1.28 (m, 2H, H-c), 1.35–1.40 (m, 2H, H-b), 1.40–1.54 (complex m, 6H, 4,6,10-adamantane H), 1.60 (dd, J=28.1, 13.0 Hz, 6H, 2,8,9-adamantane H), 1.98 (br s, 3H, 3,5,7-adamantane H), 1.97 (q, J=6.5 Hz, 1H, H-11), 2.34 (dt, J=11.4, 7.2 Hz, 1H, H-a), 2.64 (dt, J=11.4, 7.1 Hz, 1H, H-a). 13C NMR (CDCl3, 100 MHz) δ (ppm) 13.59 (C-12), 27.41 (C-c), 28.62 (3,5,7-adamantane C), 20.32 (C-b), 37.40 (4,6,10-adamantane C), 36.14 (1-adamantane C), 38.75 (2,8,9-adamantane C), 49.05 (C-a), 62.56 (C-11). ESI-MS: m/z 441.4 [M+H]+.
N,N′-bis-(1-adamantan-1-yl-ethyl)-octane-1,8-diamine (4c)
Compound 4c (90 mg, 47%) was obtained as a yellow oil from 178.93 mg of 3c. 1H NMR (CDCl3, 400 MHz) δ (ppm) 0.96 (d, J=6.5 Hz, 3H, H-12) 1.31 (m, 4H, H-c,d), 1.42–1.48 (m, 2H, H-b), 1.49–1.63 (complex m, 6H, 4,6,10-adamantane H), 1.68 (dd, J=27.1, 12.6 Hz, 6H, 2,8,9-adamantane H), 1.98 (br s, 3H, 3,5,7-adamantane H), 2.06 (q, J=6.5 Hz, 1H, H-11), 2.42 (dt, J=11.3, 7.2 Hz, 1H, H-a), 2.72 (dt, J=11.3, 7.2 Hz, 1H, H-a). 13C NMR (CDCl3, 100 MHz) δ (ppm) 13.56 (C-12), 27.41 (C-d), 29.56 (C-c), 28.61 (3,5,7-adamantane C), 30.27 (C-b), 36.14 (1-adamantane C), 37.38 (4,6,10-adamantane C), 38.73 (2,8,9-adamantane C), 49.10 (C-a), 62.61 (C-11). ESI-MS: m/z 469.38 [M+H]+.
N,N′-bis-(1-adamantan-1-yl-ethyl)-decane-1,10-diamine (4d)
Compound 4d (90 mg, 47%) was obtained as yellow oil from 188.93 mg of 3d. 1H NMR (CDCl3, 400 MHz) δ (ppm) 0.89 (d, J=6.5 Hz, 3H, H-12) 1.25–1.18 (m, 6H, H-c–e), 1.36–1.41 (m, 2H, H-b), 1.42–1.59 (complex m, 6H, 4,6,10-adamantane H), 1.6 (dd, J=24.3, 10.8 Hz, 6H, 2,8,9-adamantane H), 1.90 (br s, 3H, 3,5,7-adamantane H), 2.01 (q, J=6.5 Hz, 1H, H-11), 2.36 (dt, J=11.4, 7.3 Hz, 1H, H-a), 2.66 (dt, J=11.4, 7.2 Hz, 1H, H-a). 13C NMR (CDCl3, 100 MHz) δ (ppm) 13.43 (C-12), 27.41 (C-e) 29.51 (C-d), 29.53 (C-c), 28.59 (3,5,7-adamantane C), 30.01 (C-b), 36.14 (1-adamantane C), 37.74 (4,6,10-adamantane C), 38.76 (2,8,9-adamantane C), 49.02 (C-a), 62.68 (C-11). ESI-MS: m/z 497.46 [M+H]+.
N,N′-bis-(1-adamantan-1-yl-ethyl)-dodecane-1,12-diamine (4e)
Compound 4e (80 mg, 42%) was obtained as colorless oil from 195.03 mg of 3e. 1H NMR (CDCl3, 400 MHz) δ (ppm) 0.87 (d, J=6.5 Hz, 3H, H-12) 1.22 (m, 8H, H-c–f), 1.34–1.40 (m, 2H, H-b), 1.40–1.54 (complex m, 6H, 4,6,10-adamantane H), 1.6 (dd, J=27.8, 12.8 Hz, 6H, 2,8,9-adamantane H), 1.90 (br s, 3H, 3,5,7-adamantane H), 1.97 (q, J=6.5 Hz, 1H, H-11), 2.34 (dt, J=11.3, 7.3 Hz, 1H, H-a), 2.63 (dt, J=11.3, 7.2 Hz, 1H, H-a). 13C NMR (CDCl3, 100 MHz) δ (ppm) 13.59 (C-12), 27.46 (C-f) 29.60 (C-d,e), 29.62 (C-c), 28.62 (3,5,7-adamantane C), 30.33 (C-b), 36.14 (1-adamantane C), 37.40 (4,6,10-adamantane C), 38.74 (2,8,9-adamantane C), 49.15 (C-a), 62.60 (C-11). ESI-MS: m/z 525.48 [M+H]+.
(1-Adamantan-1-yl-ethyl)-butyl-amine (5a)
Compound 5a (70 mg, 74%) was obtained as colorless oil from 110 mg of its precursor amide. 1H NMR (CDCl3, 400 MHz) δ (ppm) 0.85 (t, J=7.3 Hz, 3H, H-d) 0.89 (d, J=6.5 Hz, 3H, H-12) 1.22–1.32 (m, 2H, H-c), 1.36–1.41 (m, 2H, H-b), 1.41–1.56 (complex m, 6H, 4,6,10-adamantane H), 1.60 (dd, J=25.2, 11.1 Hz, 6H, 2,8,9-adamantane H), 1.90 (br s, 3H, 3,5,7-adamantane H), 2.01 (q, J=6.5 Hz, 1H, H-11), 2.38 (dt, J=11.4, 7.3 Hz, 1H, H-a), 2.68 (dt, J=11.4, 7.2 Hz, 1H, H-a). 13C NMR (CDCl3, 100 MHz) δ (ppm) 13.41 (C-d) 14.02 (C-12), 20.56 (C-c), 28.59 (3,5,7-adamantane C), 32.18 (C-b), 35.98 (1-adamantane C), 37.34 (4,6,10-adamantane C), 38.68 (2,8,9-adamantane C), 48.68 (C-a), 62.69 (C-11). ESI-MS: m/z 236.16 [M+H]+.
Biochemical analysis
Generation of p7-expression constructs
The generation of genotype (GT)-specific cDNA constructs for the p7 protein of GTs 1a–4a of HCV has been described before.19
Cell culture and transfection
HEK293 cells were cultured in 10 cm tissue-culture petri dishes in MEM (Sigma-Aldrich, St Louis, MO, USA) supplemented with 10% FBS (Thermo Fisher Scientific, Waltham, MA, USA) and penicillin–streptomycin (Sigma-Aldrich) at 5% CO2 and 37°C in a water-saturated atmosphere. For experiments, cells were plated on poly-L-lysine-treated glass coverslips in 6 cm dishes and transfected 1 day after passage using 1.5 μg p7 cDNA, 1.5 μg GFP cDNA and 3 μL GenCarrier (Epoch Life Science, Sugar Land, TX, USA). Measurements were performed 2–5 days after transfection.
Electrophysiological recordings and data analysis
M2: Inhibitors were tested in a two-electrode voltage-clamp assay using Xenopus laevis frog oocytes microinjected with RNA expressing either the wild type (WT) or the S31N mutant of the A/M2 protein, as previously reported.41 The potency of the inhibitors was expressed as percentage inhibition of A/M2 current observed after 2 minutes of incubation with 100 μM compounds, and we measured inhibition as the average ± SD from three replicates.
P7: HEK293 cells, obtained from the Health Protection Agency European Cell Culture Collection (Salisbury, UK), on poly-L-lysine coated coverslips were transfected with p7 cDNA constructs 2–4 days prior to electrophysiological recordings. Current responses were measured at room temperature (21°C–23°C) at a holding potential of −60 mV using an EPC10 amplifier and Pulse software (Heka Electronics, Lambrecht, Germany). Recording pipettes were made from borosilicate glass (World Precision Instruments, Berlin, Germany) using a P-97 horizontal puller (Sutter, Novato, CA, USA). An OctaFlow system (NPI Electronics, Tamm, Germany) was used for fast perfusion of suspended single cells. The external buffer consisted of 90 mM N-methyl-D-glucamine, 3 mM CaCl2, 90 mM HEPES, and 90 mM 2-(N-morpholino) ethanesulfonic acid. The pH of the external buffer was adjusted to 8.5, 7.5, 6.5, 6.0, 5.5, 5.0 and 4.5 using NaOH or HCl. Internal buffer was 90 mM N-methyl-D-glucamine, 10 mM ethylene glycol tetraacetic acid, 180 mM HEPES, pH 7.5 (NaOH). For measurement of whole-cell p7 currents, cells were perfused continuously with recording buffer during 1-minute recording intervals. Inhibitors were diluted from stock solutions (10 mM) into pH 5.5 buffer. Inhibition measurements were started with baseline recordings of extracellular buffer pH 7.5 (4–5 seconds), followed by pH 5.5 (to induce channel opening). Then, inhibitors (at pH 5.5) were applied, followed by final control applications. Sometimes, control solution (pH 5.5 without inhibitor) was perfused between inhibitor applications. Typical current amplitudes were 100–400 pA. Dose–response curves were constructed from four to five cells per GT and inhibitor and IC50 values determined using a nonlinear fit to the equation Iobs = Imax/[1 + ([I]/IC50)], where Iobs is the observed current at any given concentration of inhibitor, Imax the maximum current amplitude observed in the absence of inhibitor, and [I] the concentration of inhibitor. IC50 for the inhibition of p7 channels was determined from the concentration-dependent inhibition of p7-mediated TM currents. Values for inhibition of GTs 1a, 2a, 3a, and 4a by amantadine and rimantadine have been previously reported.19 For this study, only a few additional measurements were performed for confirmation.
Molecular modeling
The protein was prepared using the structure preparation wizard in MOE42 (version 2015.10) and saved as a mol2 file. 3-D structures of the compounds were built and minimized using the MMFF94x force field in MOE using a gradient of 0.0001 kcal/(mol Å). Docking was performed using GOLD (version 5.3).43,44 Binding-site residues were defined by specifying the alpha carbon of Leu20 and using the default cutoff radius of 10 Å, with the “detect cavity” option enabled. GOLD docking experiments were performed using the ChemPLP scoring function. The search efficiency of the genetic algorithm was at 200% setting with the receptor kept rigid. For each compound, 50 complexes were generated and clustered based on their RMSD with the threshold set at 0.75 Å using the complete linkage method. The best-ranked pose from the most populated cluster was selected as the final pose. Figures were prepared using PyMol (version 1.8; Schrödinger, New York, NY, USA).
Results
Chemistry
The target dimeric amantadines 2a–e and rimantadines 4a–e were prepared in a two-step approach involving diamide formation with dicarboxylic acids HO2C-(CH2)n-CO2H (n=2, 4, 6, 8, 10) and subsequent reduction using borane. In the amantadine series, the dicarboxylic acids were converted to the corresponding acid chlorides using oxalyl chloride. The crude acid chlorides (1.4 mmol) were reacted with an excess of amantadine (4 mmol) to give diamides 1a–e. The target compounds 2a–e were obtained in 50%–75% yield by reduction with borane–THF complex (Figure 1).
Since rimantadine is commercially available only as a hydrochloride, an alternative and more convenient one-pot procedure for the amide formation was employed using diisopropylethylamine to release the rimantadine base from its salt and using EDCI HCl–hydroxybenzotriazole to activate the dicarboxylic acids. The rimantadine amides 3a–e were obtained in 69%–97% yield and reduced using borane–THF complex to give the target amines 4a–e in 42%–74% yield (Figure 2). For the monomeric reference compound 5a, a similar procedure was employed, where butyric acid was coupled to rimantadine followed by amide reduction using borane–THF complex.
Biological assays
M2-inhibitory activity
The inhibitory activity of all dimeric compounds was tested on the influenza virus M2 ion channels expressed in Xenopus oocytes using the two-electrode voltage-clamp technique at 100 μM concentration.29 Amantadine showed 91%±3% inhibition against WT M2 protein and was used as benchmark for M2 inhibitory activity. The newly synthesized dimeric compounds showed significantly lower inhibitory activity relative to the monomeric amantadine (Figure 3A). The compounds’ M2-blocking activity showed no dependence on alkyl-spacer length, with all dimeric compounds exhibiting relatively similar inhibition. The inhibitory activity of the reference monomeric ligand 5a (40%±5%), whose structure represents half the molecule of the rimantadine dimer 4c, was approximately fourfold that of 4c, indicating that introduction of the extra bulky adamantane group was detrimental to blocking of M2 ion-channel activity.
Examination of the experimentally determined structures of M2 protein shows an ion channel of limited pore size with its N-terminal end constricted by a hydrophobic Val27 valve. The high-resolution X-ray crystal structure of M2 protein (Protein Data Bank [PDB] 3LBW)30 shows three intercalated water clusters, which are important not only for the stability and ion-channel activity of M2 protein but also for drug binding. The upper layer of water molecules is stabilized by hydrogen bonds with the carbonyl groups of Gly34. The lower layer of water molecules lies deeper in the pore lumen, forming hydrogen bonds with His37and Trp41.
Overlaying the drug-free solid-state NMR structure of M2 protein (PDB 2KQT)31 with the amantadine-bound X-ray structure (PDB 3LBW) showed amantadine present in the pore lumen, with its adamantane cage placed in a hydrophobic groove formed mainly by Ala30and Ser31 residues (Figure 4A). When amantadine binds to the channel, it breaks the continuous water wires in the channel, which are critical for proton conductance. The positively charged ammonium group appears to mimic the conducting hydronium ion, forming hydrogen bonds with the backbone carbonyls of Gly34 that are mediated by the upper layer of water molecules. Importantly, amantadine binds to the M2-WT channel with its positively charged ammonium facing the C-termini of the channel, suggesting bulky substitutions on the amine group will not be tolerated. Our dimeric compounds (2a–e, 4a–e), with a secondary hydrophobic adamantane cage introduced in their structure, will not be expected to fit in the M2-WT channel. Indeed, as shown in Figure 3, none of the dimeric compounds had improved channel blockage against M2-WT when compared with amantadine. Our results are also in agreement with previous structure–activity relationship studies where introduction of bulky substituents on the adamantane cage drastically decreased the compounds’ M2 inhibitory activity.32 The small diameter of M2 protein pores limits the size of ligands that can fit into the pores. As a result, the second hydrophobic adamantane moiety introduced in our dimeric compounds cannot be accommodated inside the M2 pore lumen, which explains the low inhibitory activity of our dimeric compounds relative to the monomeric amantadine and ligand 5a.
  | Figure 4 Inhibition of the influenza M2 proton channel. |
The inhibitory activity of these dimeric compounds was further evaluated against S31N M2 mutant form (Figure 3B). The S31N represents the predominant amantadine-resistant M2 form.33 All the dimeric compounds showed lower inhibitory activity compared to amantadine. This could be explained by the dimeric compounds lacking the adamantane-NH2+-CH2-aryl pharmacophore of M2-S31N inhibitors.34,35 Nevertheless, the M2-inhibitory activity appears to be dependent on the alkyl-spacer length within the dimeric amantadine series: an increase in inhibitory activity was observed with increased spacer length, with the dimer linked by a ten-carbon spacer (2d) exhibiting the highest inhibitory activity.
A previous solution NMR structure of the inhibitor-bound M2-S31N structure showed that the inhibitor binds to the S31N mutant in a flipped orientation relative to that in the M2-WT channel. Specifically, the amino group from the S31N mutant-bound inhibitor is oriented upward toward the N-terminal end.36 Up till now, compounds reported to possess inhibitory activity for M2-S31N mutant form share the general adamantane-NH2+-CH2-aryl pharmacophore (Figure 4B). The positively charged ammonium group and the heteroaryl moiety from the inhibitor form water-mediated interactions with the amides from the Asn31 residues. In our dimeric compounds, this polar aryl group is missing, which explains their lack of efficacy in inhibiting the M2-S31N channel.
In summary, the lack of channel blockage of the dimeric amantadine and rimantadine analogs against M2-WT and M2-S31N mutant is consistent with previously proposed drug-binding mechanisms and further confirms that the pore-binding model is the pharmacologically relevant drug-binding model.
p7-inhibitory activity
The inhibitory activity of all compounds was tested on p7 channels for GTs 1a, 1b, 2a, 3a, and 4a expressed in HEK293 cells (Table 1). Amantadine and rimantadine are known inhibitors of p7 activity, blocking lysosomal deacidification and lowering p7 currents in patch-clamp measurements,19 and thus were used as benchmarks for comparison. The dimeric adamantanes showed similar potency to their respective monomeric analogs in GTs 1a, 1b, and 4a. Testing of the inhibitory activity of our dimeric amantadine series on GT2a showed these compounds to be at least 700-fold more potent than amantadine and eightfold more potent than rimantadine. For GT3a, the dimeric compounds were at least 150-fold more potent than amantadine and equipotent to rimantadine. A positively charged amino group appears important for binding to GT2a, as compound 2c with a secondary amine had an IC50 of 0.8 nM, whereas its corresponding amide analog 1c did not show any inhibitory activity (IC50>100 nM). On the other hand, this positively charged amine group appears less important for inhibition of other GTs, as both compounds 2c and 1c showed comparable potency in GTs 1a, 1b, 3a, and 4a.
Our results show no dependence between the p7-inhibitory activity of the dimeric compounds and alkyl-spacer length. All dimeric compounds showed relatively similar potency in the tested GTs, suggesting that all these compounds have a similar binding mode. Despite the increasing amount of structural data on p7, there is no consensus on which conformation exists during a natural infection. Because of the structural impact, the proposed binding mode of our compounds may differ depending on the 3-D model. Accordingly, we considered the two available NMR structures for p7 protein to develop hypotheses upon the binding mode of our second adamantane moiety. Two homology models for one of the GTs under study (GT2a, strain J6) were previously constructed37 using the NMR structure of HCV p7 GT5a (PDB 2M6X)26 as a template for model 1, and the NMR/MD structure of the monomeric form of GT1b as a template for model 2.38 Although the exact binding mode of amantadine–rimantadine is not clearly understood yet, it is generally agreed that they bind in a membrane-facing pocket centered on residue number 20. Structural examination of both models showed the distance between the Cα atoms of Leu20 in two adjacent adamantane-binding sites to be ~11 Å and 17 Å, respectively. The second adamantane moiety in compound 2a having the shortest spacer, cannot span this distance and thus cannot bind in the neighboring adamantane-binding site located in the adjacent monomer. This was also supported by preliminary docking studies, where the first adamantane ring in the dimeric compound 2a showed a similar binding mode as amantadine, while the second adamantane appeared protruding to another cavity (Figure 5A and B) which may suggest the presence of a secondary adamantane-binding site. Future site-directed mutagenesis would be needed to confirm unambiguously the binding mode suggested by our docking studies.
The inhibitory activity of adamantanes varied between the different GTs under study. GT-dependent sensitivity of p7 to multiple inhibitors was previously reported where amantadine could not inhibit p7 ion-channel activity in GTs 2a and 3a,17,19 similarly to what we also observed here. On the other hand, an extra ethyl group inserted between the adamantane ring and the amino group present in rimantadine led to at least 100-fold increase in potency compared to amantadine, indicating a better fit in the adamantane pocket. This could suggest that the size of the adamantane pocket in GTs 2a and 3a might be bigger than that of other GTs, and so amantadine could not induce the conformational changes necessary for favoring the closed state of p7 protein. Previous NMR and MD simulation experiments of p7 protein reported the presence of a proline residue at position 49 that forms a kink in the carboxy-terminal end of TMD2, tilting it toward TMD1 of the adjacent monomer and forming the proposed adamantane primary binding pocket.39,40 Examination of the p7 protein sequence showed both GTs 2a and 3a lacked this proline (Figure 5C). A proline residue in a TM helix typically changes the orientation of the helix, causing a kink or even breaking the helix. This common behavior results from its inability to act as a hydrogen-bond donor (tertiary amide) and steric interferences of its cyclic side chain with residues in the preceding helical turn. The lack of proline in GTs 2a and 3a could affect the kink angle of TMD2, causing possible changes in the size of the adamantane-binding pocket, which could explain the low inhibitory activity of amantadine. Our dimeric compounds; being more bulky; could overcome amantadine’s resistance in GTs 2a and 3a, showing an even higher potency than that of rimantadine.
The higher p7-inhibitory activity observed for our dimeric amantadines relative to monomeric amantadine could be attributed either to better fitting in this large adamantane pocket or simultaneous binding of the two adamantane rings in the primary and secondary pocket inducing the conformational changes necessary for favoring the closed state of p7 protein. Future point-mutation studies introducing a proline at position 49 in p7 protein in these resistant GTs should help in better understanding the binding mode of these dimeric compounds.
Conclusion
In the present study, we synthesized a series of dimeric adamantane compounds with either amantadine or rimantadine pharmacophore linked by alkyl spacers of various lengths. The antiviral activity of these compounds was studied against two viroporins: M2 and p7. Upon testing on M2 protein, the extra adamantane moiety caused a decrease in potency relative to the reference monomeric compounds amantadine and rimantadine, implying that these dimers could not be accommodated by either M2-WT or M2-S31N proton channels. These findings, although disappointing, are consistent with previously proposed ligand-binding mechanisms: ligand binds to the M2-WT with its positively charged ammonium facing the C-terminal His37, while M2-S31N inhibitors need to have the adamantane-NH2+-CH2-aryl pharmacophore and bind to the M2-S31N channel with their aryl group facing the N-terminal Val27. On the other hand, these dimers could have been lodged in the structure of p7 protein and were able to address the differences in adamantanes’ sensitivity among the various GTs of HCV. The dimeric analogs were equipotent to the monomeric reference ligands when tested on HCV GTs 1a, 1b, and 4a, while being 700-fold and 150-fold more potent than amantadine for GTs 2a and 3a, respectively. A positively charged amino group appears important for binding of amantadine to p7 channels of GT 2a, while being less important for binding to p7 of other GTs. These results will help in better understanding of the binding mode of adamantanes to the p7 protein. In summary, symmetric dimeric adamantane compounds appear to be promising p7 inhibitors across all GTs of HCV, where they may constitute a class of promising direct-acting antiviral drugs.
Acknowledgments
The authors thank Professor Ulrike Holzgrabe, University of Wuerzburg for her support. Yasmine M Mandour appreciates financial support from the German Academic Exchange Service DAAD.
Disclosure
The authors report no conflicts of interest in this work.
References
Gonzalez M, Carrasco L. Viroporins. FEBS Lett. 2003;552:28–34. | ||
Wang K, Xie S, Sun B. Viral proteins function as ion channels. Biochim Biophys Acta. 2011;1808:510–515. | ||
Henkel J, Weisz O. Influenza virus M2 protein slows traffic along the secretory pathway: pH perturbation of acidified compartments affects early Golgi transport steps. J Biol Chem. 1998;273:6518–6524. | ||
Vincent M, Raja N, Jabbar M. Human immunodeficiency virus type 1 Vpu protein induces degradation of chimeric envelope glycoproteins bearing the cytoplasmic and anchor domains of CD4: role of the cytoplasmic domain in Vpu-induced degradation in the endoplasmic reticulum. J Virol. 1993;67:5538–5549. | ||
Westerbeck JW, Machamer CE. A coronavirus E protein is present in two distinct pools with different effects on assembly and the secretory pathway. J Virol. 2015;89:9313–9323. | ||
Jones C, Murray C, Eastman D, Tassello J, Rice C. Hepatitis C virus p7 and NS2 proteins are essential for production of infectious virus. J Virol. 2007;81:8374–8383. | ||
Fischer W, Hsu H. Viral channel forming proteins: modeling the target. Biochim Biophys Acta. 2011;1808:561–571. | ||
Ouyang B, Chou J. The minimalist architectures of viroporins and their therapeutic implications. Biochim Biophys Acta. 2014;1838:1058–1067. | ||
Martin K, Helenius A. Nuclear transport of influenza virus ribonucleoproteins: the viral matrix protein (M1) promotes export and inhibits import. Cell. 1991;67:117–130. | ||
Helenius A. Unpacking the incoming influenza virus. Cell. 1992;69:577–578. | ||
Grambas S, Bennett MS, Hay AJ. Influence of amantadine resistance mutations on the pH regulatory function of the M2 protein of influenza A virus. Virology. 191:541–549. | ||
Steinmann E, Penin F, Kallis S, Patel A, Bartenschlager R, Pietschmann T. Hepatitis C virus p7 protein is crucial for assembly and release of infectious virions. PLoS Pathog. 2007;3:e103. | ||
Farag NS, Breitinger U, El-Azizi M, Breitinger HG. The p7 viroporin of the hepatitis C virus contributes to liver inflammation by stimulating production of interleukin-1β. Biochim Biophys Acta. 2017;1863:712–720. | ||
Davies WL, Grunert RR, Haff RF, et al. Antiviral activity of 1-adamantaneamine (amantadine). Science. 1964;144:862–863. | ||
Pinto LH, Holsinger LJ, Lamb RA. Cell influenza virus M2 protein has ion channel activity. Cell. 1992;69:517–528. | ||
Wang C, Takeuchi K, Pinto LH, Lamb RA. Ion channel activity of influenza A virus M2 protein: characterization of the amantadine block. J Virol. 1993;67:5585–5594. | ||
Griffin S, Stgelais C, Owsianka AM, Patel AH, Rowlands D, Harris M. Genotype-dependent sensitivity of hepatitis C virus to inhibitors of the p7 ion channel. Hepatology. 2008;48:1779–1790. | ||
Griffin S, Beales LP, Clarke DS, et al. The p7 protein of hepatitis C virus forms an ion channel that is blocked by the antiviral drug, amantadine. FEBS Lett. 2003;535:34–38. | ||
Breitinger U, Farag NS, Ali NK, Breitinger HG. Patch-clamp study of hepatitis C p7 channels reveals genotype-specific sensitivity to inhibitors. Biophys J. 2016;110:2419–2429. | ||
Schnell JR, Chou JJ. Structure and mechanism of the M2 proton channel of influenza A virus. Nature. 2008;451:591–595. | ||
Stouffer AL, Acharya R, Salom D, et al. Structural basis for the function and inhibition of an influenza virus proton channel. Nature. 2008;451:596–599. | ||
Wang J, Li F, Ma C. Recent progress in designing inhibitors that target the drug-resistant M2 proton channels from the influenza A viruses. Biopolymers. 2015;104:291–309. | ||
Pielak RM, Oxenoid K, Chou JJ. Structural investigation of rimantadine inhibition of the AM2-BM2 chimera channel of influenza viruses. Structure. 2011;19:1655–1663. | ||
Cook GA, Opella SJ. NMR studies of p7 protein from hepatitis C virus. Eur Biophys J. 2010;39:1097–1104. | ||
Patargias G, Zitzmann N, Dwek R, Fischer WB. Protein-protein interactions: modeling the hepatitis C virus ion channel p7. J Med Chem. 2006;49:648–655. | ||
OuYang B, Xie S, Berardi MJ, et al. Unusual architecture of the p7 channel from hepatitis C virus. Nature. 2013;498:521–525. | ||
Foster TL, Thompson GS, Kalverda AP, et al. Structure-guided design affirms inhibitors of hepatitis C virus p7 as a viable class of antivirals targeting virion release. Hepatology. 2014;59:408–422. | ||
Sokolova AS, Yarovaya OC, Korchagina DV, et al. Camphor-based symmetric diimines as inhibitors of influenza virus reproduction. Bioorg Med Chem. 2014;22:2141–2148. | ||
Li F, Hu Y, Wang Y, Ma C, Wang J. Expeditious lead optimization of isoxazole-containing influenza A virus M2-S31N inhibitors using the Suzuki–Miyaura cross-coupling reaction. J Med Chem. 2017;60:1580–1590. | ||
Acharya R, Carnevale V, Fiorin G, et al. Structure and mechanism of proton transport through the transmembrane tetrameric M2 protein bundle of the influenza A virus. Proc Natl Acad Sci U S A. 2010;107:15075–15080. | ||
Cady SD, Schmidt-Rohr K, Wang J, Soto CS, Degrado WF, Hong M. Structure of the amantadine binding site of influenza M2 proton channels in lipid bilayers. Nature. 2010;463:689–692. | ||
Duque MD, Ma C, Torres E, et al. Exploring the size limit of templates for inhibitors of the M2 ion channel of influenza A virus. J Med Chem. 2011;54:2646–2657. | ||
Furuse Y, Suzuki A, Oshitani H. Large-scale sequence analysis of M gene of influenza A viruses from different species: mechanisms for emergence and spread of amantadine resistance. Antimicrob Agents Chemother. 2009;53:4457–4463. | ||
Li F, Ma C, DeGrado WF, Wang J. Discovery of highly potent inhibitors targeting the predominant drug-resistant S31N mutant of the influenza A virus M2 proton channel. J Med Chem. 2016;59:1207–1216. | ||
Li F, Ma C, Hu Y, Wang Y, Wang J. Discovery of potent antivirals against amantadine-resistant influenza A viruses by targeting the M2-S31N proton channel. ACS Infect Dis. 2016;2:726–733. | ||
Wu Y, Canturk B, Jo H, et al. Flipping in the pore: discovery of dual inhibitors that bind in different orientations to the wild-type versus the amantadine-resistant S31N mutant of the influenza A virus M2 proton channel. J Am Chem Soc. 2014;136:17987–17995. | ||
Montserret R, Saint N, Vanbelle C, et al. NMR structure and ion channel activity of the p7 protein from hepatitis C virus. J Biol Chem. 2010;285:31446–31461. | ||
Scull MA, Schneider WM, Flatley BR, et al. The N-terminal helical region of the hepatitis C virus p7 ion channel protein Is critical for infectious virus production. PLoS Pathog. 2015;11:e1005297. | ||
Cook GA, Dawson LA, Tian Y, Opella SJ. Three-dimensional structure and interaction studies of hepatitis C virus p7 in 1,2-dihexanoyl-sn-glycero-3-phosphocholine by solution nuclear magnetic resonance. Biochemistry. 2013;52:5295–5303. | ||
Chandler DE, Penin F, Schulten K, Chipot C. The p7 protein of hepatitis C virus forms structurally plastic, minimalist ion channels. PLoS Comput Biol. 2012;8:e1002702. | ||
Rey-Carrizo M, Torres E, Ma C, et al. 3-Azatetracyclo[5.2.1.1(5,8).0(1,5)]undecane derivatives: from wild-type inhibitors of the M2 ion channel of influenza A virus to derivatives with potent activity against the V27A mutant. J Med Chem. 2013;56:9265–9274. | ||
Chemical Computing Group. Molecular Operating Environment version 2013.08. 2017. Available from: https://www.chemcomp.com/MOE-Molecular_Operating_Environment.htm. Accessed March 19, 2018. | ||
Jones G, Willett P, Glen RC. Molecular recognition of receptor sites using a genetic algorithm with a description of desolvation. J Mol Biol. 1995;245:43–53. | ||
Jones G, Willett P, Glen RC, Leach AR, Taylor R. Development and validation of a genetic algorithm for flexible docking. J Mol Biol. 1997;267:727–748. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.