Back to Journals » OncoTargets and Therapy » Volume 17
SIM2: Its Prognostic Significance and Oncogenic Role in Endometrial Carcinoma
Authors Wei Y, Zhao X, Tang H, Ma J, Wang Y, Li L
Received 18 September 2023
Accepted for publication 9 January 2024
Published 26 January 2024 Volume 2024:17 Pages 45—61
DOI https://doi.org/10.2147/OTT.S440788
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr John Riches
Yunfang Wei,* Xianlei Zhao,* Hong Tang, Jin Ma, Yongfeng Wang, Linxia Li
Department of Obstetrics & Gynecology, Shanghai Seventh People’s Hospital Affiliated to Shanghai University of Traditional Chinese Medicine, Shanghai, 200137, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Linxia Li, Email [email protected]
Background: Endometrial carcinoma ranks as the second most widespread malignancy affecting the reproductive system in females. Effective prognostic biomarkers are required to further improve survival rates for patients. Single-minded homolog 2 (SIM2) is known to participate in neurogenesis as a transcription factor. However, the potential role of SIM2 in endometrial carcinoma remains elusive.
Methods: Multiple public databases, including TIMER2.0, GEIPA2, UALCAN, LinkedOmics, BioGRID, DAVID and cBioPortal, were used to investigate SIM2 mRNA expression, SIM2-associated genes, PPI network, functional enrichment analysis, SIM2 gene alterations and methylation. The association between SIM2 expression and immune cell infiltrates was explored using GSVA. The effects of gene alterations and methylation on patient survival and CD8+T infiltration were examined using GSCA. Moreover, the prognostic potential of SIM2 was evaluated using COX regression, ROC curves and a nomogram model. Finally, the differential expression and function of SIM2 in UCEC were explored using qPCR, WB, CCK8 and Transwell assays.
Results: Our findings revealed the heightened expression of SIM2 in endometrial carcinoma, and that its DNA methylation and CNV alterations were correlated with immune infiltration and patients’ prognosis. Additionally, functional enrichment revealed the involvement of SIM2 in transcription regulation and signal transduction. Moreover, we performed cell-based experiments to corroborate the oncogenic function of SIM2 in facilitating cell proliferation, migration and invasion.
Conclusion: Collectively, these results suggest that SIM2 holds promise as both a potential prognostic indicator and a viable treatment target for endometrial carcinoma.
Keywords: uterine corpus endometrial carcinoma, DNA methylation, genetic alterations, immune cell infiltration, prognosis
Introduction
Uterine Corpus Endometrial Cancer (UCEC) is a type of neoplasm primarily afflicting peri- and post-menopausal women, typically found localized within the endometrium.1 In 2020, there were approximately 380,000 novel cases of UCEC reported worldwide, making it the second most common malignancy in the female reproductive system, ranking just after cervical cancer.1 Historically, UCEC displayed a favorable prognosis, characterized by elevated cure rates and an overall 5-year survival rate ranging between 75% and 80% following surgical intervention.2 Despite numerous attempts to improve post-operative prognosis, chemoresistance remains a challenge, especially for patients at advanced stages.3,4 Furthermore, over the past 40 years, the incidence of UCEC has doubled, which has been attributed to high-risk factors such as obesity, diabetes, estrogen use and polycystic ovary syndrome.5 Consequently, exploring new biomarkers and therapeutic targets for UCEC has become particularly important for clinical practice and research.
SIM2 (Single-minded homolog 2) is a homolog of Drosophila simple-minded gene, encoding a master transcription factor.6 As SIM2 can form complexes with aryl hydrocarbon receptor nuclear translocator (ARNT) or ARNT2, its role in multiple biological processes, such as neurogenesis and angiogenesis, has been demonstrated.7 A dimer comprising SIM2 and ARNT2 specifically binds to central midline elements (CME) or hypoxia-response elements, thus orchestrating gene expression regulation. In addition, accumulating evidence has underscored SIM2’s involvement in the etiology of various cancer types. For instance, SIM2 downregulates SNAI2 in breast cancer, thus repressing epithelial–mesenchymal transition and impeding tumor growth and invasion.8 Conversely, SIM2 knockdown impedes tumor growth and triggers apoptosis in colon carcinoma and pancreatic carcinoma.9,10 Furthermore, SIM2 has been identified as a biomarker for prostate cancer.11,12 Overall, the role of SIM2 is disease-dependent, and its function in UCEC has remained elusive.
Here, we utilized composite online bioinformatic tools to comprehensively investigate clinical and biological attributes of SIM2 regarding UCEC, including differential expression, associated genes, functional enrichment, gene alterations and immune cell infiltration. In parallel, we constructed a nomogram harnessing both univariate and multivariate Cox regression in an effort to assess the diagnostic potential of SIM2. Finally, cell-based experiments corroborated our findings that SIM2 acts as an oncogene. Collectively, our findings indicate that SIM2 bears clinical relevance in the pathogenesis of UCEC, and it holds potential for predicting UCEC diagnosis and prognosis, thus offering crucial insights for clinical management of UCEC.
Methods and Materials
Screening of Prognostic Genes in UCEC
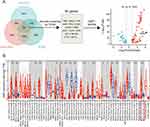
To identify differentially expressed genes predictive of survival status in UCEC patients, we analyzed gene expression data from TCGA, GSE3013 and GSE17025 datasets. A log fold change (logFC) greater than 1 and an adjusted p-value less than 0.05 were utilized as the cutoff criteria for differential expression between tumor and control samples. By overlapping differentially expressed genes, we discovered 404 common genes from these three datasets. Subsequently, a secondary screening was conducted by excluding genes with insignificant prognostic relevance based on TCGA data. Finally, 80 genes were retained and ranked according to their logFC and adjusted p-value. Among them, SIM2 showed the most heightened expression in UCEC and has not been reported in its carcinogenesis yet. In addition, SIM2 expression between cancerous and non-cancerous tissues across diverse types of cancer was explored with TIMER2.0.
SIM2 Expression in UCEC
We conducted a transcriptomic analysis on a dataset comprising 554 tumor samples and 35 normal samples from TCGA. R software was applied to decipher RNA-seq data and clinical information. We utilized the “stats”, “car” and “ggplot2” packages to explore relative SIM2 expressions in paired and non-paired normal tissue and UCEC tumor samples. Meanwhile, we explored SIM2 expression across various sample types, stages and menopause status using UALCAN. Immunohistochemical (IHC) images of SIM2 in the endometrium were acquired from the Human Protein Atlas (HPA).13
SIM2-Associated Genes and Functional Analysis
Using LinkedOmics, we searched for and visualized SIM2-associated genes in a volcano plot. Additionally, we provided a list of the top 50 genes that showed positive or negative associations. The foremost 50 genes displaying either positive or negative associations were also presented. Enrichment analysis of SIM2 was executed using DAVID to investigate the biological significance underpinning the gene lists. Specifically, we uploaded the top 50 associated genes to DAVID, and subsequently conducted functional enrichment based on the REACTOME datasets. A comprehensive exploration of the biological processes (BP), molecular function (MF) and cell components (CC) of the associated genes was conducted with GO enrichment analysis. The results were visualized in bar plots generated with GraphPad Prism 8.
Protein Interaction Networks
Using BioGrid, an online biomedical interaction repository that contains protein–protein interaction data from low- and high-throughput sequencing,14 we generated protein–protein interaction (PPI) networks centered on SIM2. Proteins that interacted physically or functionally with SIM2 were subsequently subjected to REACTOME and GO analysis with DAVID. Finally, the correlation of each gene with SIM2 was examined by TIMER2.0.
SIM2 DNA Methylation and Its Significance
With UALCAN, promoter methylation level of SIM2 was explored using data from 46 normal and 438 UCEC samples. The association between SIM2 expression and patient prognosis and DNA methylation was explored with GSCA (Gene Set Cancer Analysis), a platform for oncogene analysis.
SIM2 Gene Mutation Characterization
cBioPortal was applied to analyze SIM2 mutation and copy number alterations (CNV) in different UCEC studies. Additionally, the frequencies of SIM2 CNV events and their correlation with patient survival were explored with GSCA.
SIM2 and Immune Microenvironments
To evaluate the influence of SIM2 on immune cell infiltration, we applied R package “GSVA” to analyze the dependence of 24 tumor-infiltrating lymphocytes on SIM2.15 The stromal, immune and estimate score—which together reflect the presence of stromal and infiltrating immune cells—were computed using the ESTIMATE method.15 Besides, TIMER2.0 was used to elucidate the correlation between SIM2 mutation and infiltrated CD8+ T cell, sCNA (somatic copy number alterations) status and CD8+ T infiltrates regarding UCEC patients’ survival. Finally, we scrutinized the association between SIM2 methylation and the presence of CD8, Tfh (T follicular helper), Cytotoxic and iTreg (induced T regulatory) through the use of GSCA.
Univariate and Multivariate Cox Regression
We used R packages “survival[3.3.1]” and “rms[6.3–0]” for univariate analysis to scrutinize correlations between OS (overall survival) and prognosis-related variables encompassing clinical stage, age, histological type, tumor invasion and SIM2 expression. Certain variables with a p-value less than 0.02 were singled out for inclusion in the creation of a multi-factor Cox model, from which two forest plots were generated. Furthermore, the nomogram and the calibration curve were also visualized to estimate survival using independent risk factors of multivariate analysis. In addition, we explored and visualized the prediction efficiency of our nomogram model with the R package “timeROC[0.4]” and “ggplot2[3.3.6]”.
SIM2 and Patient Survival Analysis
R packages “survival”, “survminer” and “ggplot2” were used to analyze and visualize the effect of SIM2 expression on UCEC prognosis using Kaplan-Meier. P-value and hazard ratio (HR) were analyzed. Additionally, an ROC (receiver operating characteristic) curve was created to illustrate the diagnostic effectiveness of SIM2.
Cell Culture and Transfection
ISHIKAWA and HEC-1A, two endometrial cancer cell lines obtained from Procell, were maintained in DMEM (Hyclone) under 5% CO2 at 37°C. SIM2 siRNA was transfected into ISHIKAWA and HEC-1A using Lipofectamine 3000 (ThermoFisher) as instructed. Briefly, when the cells reached a confluence over 50%, cell transfection was performed using a mixture of Lipofectamine 3000 and SIM2 siRNA at a ratio of 1/20. The cells were incubated for 24 hours before downstream analysis. The siRNA sequence targeting SIM2 was GCCTTGTCTACCTCACAAGAA. Cells transfected with si-negative control oligos were considered as a negative control group.
Western Blot Analysis
In accordance with the Ethics Committee of the Seventh People’s Hospital of Shanghai University of Traditional Chinese Medicine and the Declaration of Helsinki, clinical endometrial tissues were collected with informed consent from patients and stored in the liquid nitrogen. The tissues were lysed in RIPA buffer (Beyotime), supplemented with the proteinase inhibitor cocktail (Roche). Then, lysates were extracted, quantified with a BCA protein assay kit (ThermoFisher) and subsequently separated using 10% SDS-PAGE and transferred onto PVDF membranes (Roche). After blockage with 3% non-fat milk dissolved in PBS for 1 hour at room temperature, membranes were co-incubated with primary antibodies. Membranes underwent three rounds of washing prior to being incubated with horseradish (HRP)-conjugated secondary antibodies for 1 hour at room temperature. The blots were visualized using ChemiScope (ClinX). GAPDH was used as the internal control. The following antibodies were used: mouse anti-GAPDH (1:1000, ab8245, Abcam), and rabbit anti-SIM2 (1:1000, ab131161, Abcam), Goat Anti-Mouse IgG (H&L) [HRP] (1:10,000, A00160, GenScript) and HRP-conjugated Affinipure Goat Anti-Rabbit IgG (H+L) (1:10,000, SA00001-2, Proteintech).
RT-qPCR
ISHIKAWA and HEC-1A total RNA were isolated using AG RNAex Pro (Accurate Biology). Reverse transcription of RNA was completed with HiScript RT SuperMix (Vazyme). The cDNA was stored at −80°C for later use. RT-qPCR was performed using ChamQ SYBR qPCR Master Mix (Vazyme). GAPDH served as internal control. Primer sequences for SIM2 were as follows: F: AAGGAAAATGGCGAGTTTTACGA; R: CGCGTCTCCTAAACCTTCGG. The primer sequences for GAPDH were as follows: F: ACAACTTTGGTATCGTGGAAGG; R: GCCATCACGCCACAGTTTC.
Cell Proliferation Assay
Cells were suspended at 1 × 104 cells/mL, respectively. Two hundred microliters medium containing ISHIKAWA or HEC-1A was added into 96-well plates and cultured under 5% CO2 at 37°C. Ten microliters CCK-8 solution (Beyotime) was pipetted into each chamber and incubated for 30 min before absorbance measurement. The absorbance was recorded with a microplate reader at 450nm.
Transwell Migration and Invasion Assay
For migration assays, 300 μL DMEM was added into each well in the 24-well plate. Transwell chambers (Falcon) were lowered into the 24-well plate, and 500 μL medium containing ISHIKAWA or HEC-1A cells (1 × 104 cells/mL) was gently introduced into the upper chamber carefully. Following a 24-hour incubation period under 5% CO2 at 37°C, medium in the upper chamber was discarded and the cells were meticulously disposed. Once fixed, cells at the bottom of the chamber underwent crystal violet staining for 10 min. Then, extra dye was removed with a cotton swab. The chamber was photographed with a Nikon microscope. For invasion assays, the Transwell chambers were prepared with Matrigel (Corning), with a working concentration of 50 mg/L.
Statistical Analysis
Statistical analysis was conducted with RStudio and plots were generated with GraphPad Prism 8 and RStudio, except for results obtained from public databases. Significance was defined as p < 0.05.
Results
Screening of Prognostic Markers in UCEC
Figure 1A illustrates the screening of SIM2 in terms of differential expression and prognosis in UCEC patients. In brief, 404 genes were identified from 3 transcriptome data sources: TCGA, GSE3013 and GSE17025. Among them, 80 genes were found to be prognostically relevant to UCEC. SIM2 was chosen for further analysis due to its highest logFC. Subsequently, SIM2 expression between tumor and adjacent non-tumor was explored with TIMER2.0. The analysis unveiled a significant up-regulation of SIM2 across various cancer types, including UCEC (Figure 1B).
Expression and Clinical Significance of SIM2 in UCEC
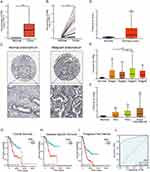
To ascertain the expression of SIM2 in UCEC, we obtained the UCEC-related transcriptome and patient clinical data from TCGA. Our analysis revealed a significant upregulation of SIM2 in malignant UCEC tissues, as depicted in Figure 2A and B. Furthermore, we corroborated these findings by examining SIM2 protein expression through HPA, which confirmed its elevated levels compared to normal tissues (Figure 2C). Additionally, we utilized UALCAN to validate the heightened expression of SIM2 in UCEC (Figure 2D). The clinical and genetic attributes of UCEC patients are comprehensively summarized in Table 1. Notably, SIM2 expression was found positively correlated with histological grade, tumor invasion and menopause status. Additionally, significant differences in SIM2 expression were found between stage I and III, as well as between stage II and III (Figure 2E). Furthermore, a correlation was observed between SIM2 expression and menopause status, with post-menopausal patients exhibiting higher SIM2 expression (p < 0.001) (Figure 2F). Subsequently, Kaplan–Meier analysis was conducted on SIM2 in UCEC patients, revealing that heightened SIM2 expression was unfavorable to patient prognosis, regarding overall survival (OS) and disease-specific survival (DSS) and progression-free interval (PFI) (Figure 2G-I). ROC analysis reflects the diagnostic potential of a certain factor in regard to sensitivity and specificity.16 With an area under the curve (AUC) of 0.914, Figure 2J demonstrates SIM2’s excellent accuracy in diagnosis. Collectively, these findings suggest that SIM2 serves as an oncogene in UCEC, highlighting its diagnostic potential.
 |
Table 1 Association Between SIM2 Expression and Clinicopathologic Features in UCEC |
Co-Expressed Genes of SIM2 and Functional Enrichment
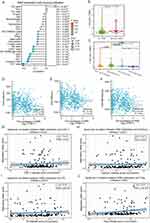
To gain biological insight into SIM2, the LinkFinder feature of LinkedOmics was employed to discover genes that are co-expressed with SIM2. In Figure 3A, the volcano plot displayed genes positively associated with SIM2 represented by red dots, while green dots represented genes negatively associated. Then, the 50 genes with the highest positive or negative association were displayed in two heatmaps (Figure 3B and C). Subsequently, 50 genes with the strongest association were chosen for functional enrichment analysis by DAVID. Biological process (BP) enrichment analysis uncovered the involvement of SIM2-associated genes in signal transduction, positive regulation of transcription from the RNA polymerase II promoter and positive regulation of transcription, while cellular compartment (CC) enrichment analysis suggested the presence of co-expressed proteins in endoplasmic reticulum membrane, chromatin and cell surface (Figure 3D). Molecular function (MF) enrichment analysis indicated the associated genes’ role in DNA binding, RNA polymerase II transcription factor activity and transcription factor activity (Figure 3D). Furthermore, the REACTOME pathway enrichment result unveiled the involvement of SIM2 in Cytokine Signaling in Immune System, Interferon Gamma Signaling, Interferon Signaling, Toll-Like Receptor 4(TLR4) Cascade and Toll-like Receptor Cascades (Figure 3E). Accordingly, the Gene Set Enrichment Analysis (GSEA) of SIM2-associated genes revealed their negative correlation with numerous immune processes, such as Interferon Signaling, Interferon Gamma Signaling, Cytokine Signaling in Immune System, Innate Immune System and TCR Signaling, whereas positively correlated with GPCR Ligand Binding (Figure 3F).
Interactive Protein Network of SIM2
We applied the BioGRID database to visualize a network of proteins that engage in physical interactions with SIM2 (Figure 4A). Next, the genes encoding these interacting proteins underwent functional enrichment analysis, which indicated a significant enrichment in regulation of transcription from the RNA polymerase II promoter, positive regulation of transcription and positive regulation of transcription from the RNA polymerase II promoter for BP, nucleus, nucleoplasm and chromatin for CC, and protein binding, RNA polymerase II transcription factor activity and DNA binding for MF (Figure 4B). Additionally, these genes were correlated with gene expression (Transcription), cellular responses to stress and stimuli (Figure 4C). Finally, we examined the correlation of each gene with SIM2 using TIMER2.0, and the results indicated significantly positive correlations between SIM2 and CIC, ARIH1, BRCA1, MAD2L2, FOXN1 and HSP90AA1 (Figure 4D).
SIM2 Promoter Methylation and Its Clinical Significance
To explore epigenetic regulation of SIM2, the promoter methylation status of SIM2 in UCEC was uncovered by UALCAN. Strikingly, tumor tissues showed an obviously higher SIM2 methylation status (Figure 5A). Additionally, GSCA was employed to examine the relationship between SIM2 methylation and expression. The result revealed that a higher methylation status indicated a lower SIM2 mRNA expression (Cor=−0.5, p < 0.05) (Figure 5B). To explore whether SIM2 methylation affects UCEC patients’ survival, we harnessed the module of Methylation & Survival in GSCA. The findings distinctly indicated an inverse relationship, where increased SIM2 DNA methylation was linked to worse OS, DSS and PFS (progression-free survival) (p = 0.01, 0.026 and 0.017, respectively) (Figure 5C-E).
Genetic Alterations of SIM2 and Its Clinical Significance
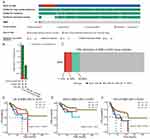
Given that genetic mutations are indicative of unfavorable cancer prognosis,17 we proceeded to scrutinize SIM2 genetic alterations in UCEC samples based on the cBioPortal database. In brief, we carried out profiling to investigate the presence of copy number alterations, mutations and structural variants in the SIM2 gene locus across 610 samples from 2 UCEC studies Endometrial Carcinoma (CPTAC, Cell 2020) and Uterine Corpus Endometrial Carcinoma (TCGA, PanCancer Atlas). In total, the frequency of SIM2 alterations was 2.8% of all samples, among which 2 samples exhibited amplification, 1 sample displayed truncating mutation, 1 sample harbored splice mutation and 13 samples featured missense mutations (Figure 6A). The distribution of SIM2 mutation and amplification frequencies across different datasets is illustrated in Figure 6B. Subsequently, we examined SIM2 CNV in UCEC using GSCA. Out of all the UCEC samples analyzed, 9.28% demonstrated amplification-comprising 8.16% with heterogenous amplification and 1.11% with homologous amplification. Moreover, a total of 10.02% cases exhibited deletions, all of which were heterogenous (Figure 6C). Finally, we delved into the relationship between patient survival and SIM2 CNV status. As illustrated in Figure 6D-F, both amplification and deletion of SIM2 correlated with worse OS, DSS and PFS outcomes.
Correlation Between SIM2 and Immune Microenvironment
As tumor progression and prognosis are closely linked to immune microenvironments, we investigated how SIM2 was correlated with the infiltration of various immune cells. Our findings indicated that SIM2 was negatively associated with Th17, Cytotoxic, pDC, iDC, T, CD8+T and NK (Figure 7A). Additionally, Stromal, Immune and ESTIMATE scores, which bear implications for prognosis, were negatively correlated with SIM2 (Figure 7D-F). It is worth noting that CD8+ T cell abundance has been demonstrated to reflect a better prognosis across a range of cancer types.18 Based on previous findings, we extended our investigation to explore the association between SIM2 alterations and CD8+T cell infiltrates. The results, as depicted in Figure 7B and C, illustrated notable variation in CD8+ T cell infiltration scores between wild-type (WT) SIM2 and mutated SIM2 groups (p = 0.013), in which samples with SIM2 amplification suggested the lowest CD8+T cell infiltration (p = 0.0068). Additionally, we conducted an analysis to examine how SIM2 methylation status is linked to the abundance of immune cells, based on ImmuCellAI from the GSCA database. Figure 7G-J illustrates the positive association between CD8+T, Cytotoxic, Tfh and iTreg cells and SIM2 methylation, indicating that SIM2 methylation may regulate immune cell infiltration.
Analysis of Prognostic Factors in UCEC
We conducted an in-depth exploration of the independent prognostic factors affecting UCEC patients’ OS. Outcomes from the univariate analysis unveiled several OS-associated factors, including high expression of SIM2, Stage III and IV, Age >60, Mixed and Serous tumor type and ≥50% tumor invasion (Figure 8A). These risk factors were further included in the multivariate analysis. As depicted in Figure 8B, SIM2 high expression, Stage III and IV, mixed tumor type and ≥50% tumor invasion were affirmed as independent risk factors. Based on these findings, we proceeded to generate a nomogram capable of estimating longevity outcomes at 1-, 3- and 5-year time points, which was validated using a calibration curve (Figure 8C and D). The nomogram exhibited promising predictive potential for UCEC patient outcomes, as demonstrated by the ROC analysis plot (Figure 8E).
Expression and Functional Validation of SIM2 in UCEC
To further confirm the involvement of SIM2 in UCEC, we gathered samples of both normal and UCEC endometrium and assessed the protein expression of SIM2. Figure 9A indicates a higher expression of SIM2 in endometrial cancer, thereby corroborating the observations drawn from our preceding bioinformatic analyses. Next, we knocked down SIM2 expression in ISHIKAWA and HEC-1A with siRNA to investigate its function in endometrial carcinogenesis. The knockdown efficiency is demonstrated in Figure 9B-C. The CCK-8 analysis demonstrated that the suppression of SIM2 significantly hindered cell proliferation in both cell lines (Figure 9D and E). Additionally, the inhibition of SIM2 exerted a marked reduction in cell motility, as evidenced by migration and invasion assays (Figure 9F and K). Overall, these in vitro experiments underscored the oncogenic role of SIM2 in UCEC.
Discussion
In recent years, advancements in diagnostic and classification tools have significantly impacted the management of endometrial cancer, ushering in a new era of personalized treatment. Based on genomic-level characterization, endometrial cancer can be further classified to achieve risk stratification and targeted therapies.19 Notably, patients with microsatellite instability (MSI), which is attributed to genetically deficient DNA mismatch repair capability, demonstrated an enhanced response to Pembrolizumab, a PD1 mono-antibody, and a more favorable prognosis.20 Therefore, the need for more refined diagnosis and classification underscores the necessity to discover new and stable molecular markers. On the other hand, the integration of medical imaging and artificial intelligence has facilitated the high-throughput analysis of quantitative information, known as radiomics.21 This approach has produced exciting findings. For example, three-dimensional preoperative MRI-based radiomics can distinguish patients from low- to high-risk groups according to histopathological features.22 Moreover, the synergy between ultrasonographic features and molecular profiling has been explored, leading to the identification of novel prognosis biomarkers from discordant data.21 Collectively, the ongoing pursuit of early diagnosis and refined classification, coupled with novel biomarkers and the development of novel techniques contributes to a more personalized and precise medical care in the realm of endometrial cancer.
Aberrant expression of SIM2 has been intimately connected to the progression of several cancer types. For instance, SIM2 over-expression has been established in prostate cancer,23,24 where it was reported to be associated with cancer aggressiveness and affect chromatin accessibility at androgen-receptor binding sites.12 Conversely, diverse research has showcased SIM2’s tumor suppressive role in breast cancer.8 Specifically, its shorter splicing variant, SIM2-s, was observed to be downregulated, consequently promoting a more invasive phenotype.25 Additionally, in cervical carcinoma, the long isoform of SIM2 (SIM2-l) was found to suppress HIF-1α-mediated angiogenesis and impede tumor growth both in vivo and in vitro.7 Nevertheless, how SIM2 contributes to UCEC pathogenesis has not yet been elucidated. In this study, we unveiled the heightened expression of SIM2 in UCEC tissues. Particularly, SIM2 expression displayed relevance to UCEC cancer stages and patients’ menopause status, inversely correlating with OS, DSS and PFI. Then, a nomogram was generated for patient survival prediction based on uni- and multi-variate Cox regression, in which SIM2 high expression, clinical stage, histological type and cancer invasion were considered independent risk factors in UCEC. Following bioinformatic investigations, we performed cell-based experiments to scrutinize the function of SIM2 in endometrial cancer cells. Our findings implied that SIM2 may function as an oncogene and hold promise as a potential prognostic biomarker for UCEC.
SIM2, belonging to the bHLH-PAS family of transcription factors, is recognized as a major contributor to Down’s syndrome.26 Similar to HIF and aryl hydrocarbon receptor (AHR), two known bHLH-PAS proteins, SIM2 engages in the formation of a dimer with ARNT or ARNT2. This complex subsequently attaches to the CME of gene regulatory regions, and suppresses transcription through SIM2’s carboxy-terminal trans-repression domain.27 Additionally, SIM2 competes for DNA elements with other bHLH-PAS proteins such as HIF-1α, thus impinging on multiple carcinogenic pathways when dysregulated.27 However, the specific role of SIM2 in UCEC is unknown. Here, we revealed that the underlying mechanism through which SIM2 exerts its effect might be related to signal transduction and transcription regulation, as identified by GO and REACTOME analysis. Furthermore, we constructed a PPI network encompassing proteins physically associated with SIM2, which again confirmed its role in signal transduction and transcription regulation.
Genetic alterations, including mutations, deletions or copy number variance of gene segments, can affect gene expression and predispose to cancers.28 Here, we investigated the genetic alteration profiles of SIM2 on cBioPortal and uncovered missense mutation and amplification of SIM2 as the predominant genetic alterations in UCEC samples. Notably, SIM2 CNV emerged as linked to OS, DSS and PFS in patients afflicted with UCEC. On the other hand, DNA methylation affects gene expression without altering nucleotide sequences.29 Aberrant DNA methylation events have been reported for numerous tumor suppressor genes and oncogenes.30 For instance, significant contributions of DNA methylation have been noted for genes such as PTEN,31 RASSF1A32 and BMP33 in the context of endometrial carcinogenesis. Here, we uncovered that SIM2 methylation was inversely associated with its mRNA expression, despite that SIM2 promoter methylation level was higher in UCEC samples. Furthermore, we discovered that SIM2 methylation was clinically relevant, as lower SIM2 methylation levels were indicative of worse OS, DSS and PFS in UCEC patients.
Tumor progression and prognosis are closely linked to the immune microenvironment, and UCEC has been extensively studied for its aberrant immune infiltration patterns.34,35 Here, SIM2 exhibited a negative association with Th17, Cytotoxic, DC, CD8+T and NK, while it displayed a positive association with Th2. As an orchestrator in autoimmune disease and infection,36 Th17’s role in cancer is controversial and context-dependent, in that Th17 cells releasing Il-22 upon the activation of TGF-βsignaling contribute to colitis-associated colon cancer,37 whereas in breast cancer increased IL-17 predicted better patient outcome.38 Here, the significant negative correlation between SIM2 and Th17 infiltrates is noteworthy, but the underlying mechanism and the impact on the tumor microenvironment were undisclosed, warranting further investigation. In addition, CD8+T infiltration has been considered as a predictor of favorable outcomes for cancer patients.39–41 Recently, it has been reported that PD-1hi, CD39, CXCL13 and CD103 CD8 positive tumor infiltrating cells are indicative of prolonged survival of UCEC patients.42 Additionally, gene mutations frequently exert influence on the immune microenvironment.43 Similar observations have been made for DNA replicase (POLE): POLE-mutated endometrial cancer frequently displayed strikingly high tumor infiltrating lymphocytes intensity, which in part explains its excellent prognosis.44 Therefore, we speculated that SIM2 gene mutation might likewise influence the anti-tumor immune response. Here, we observed a negative relationship between CD8+T infiltration and SIM2 mutation, amplification in particular, which further verified the immunosuppressive function of SIM2 in UCEC. Finally, the association between immune infiltrates and SIM2 methylation was also investigated. Our findings unveiled a positive correlation between SIM2 methylation with CD8+T, Cytotoxic, Tfh and iTreg cell infiltration. Collectively, our findings suggest that elevated SIM2 expression indicates an immune suppressive microenvironment in UCEC.
Conclusion
In summary, our study proposes that SIM2 may exert oncogenic functions and serve as a potential diagnostic and prognostic marker for UCEC, supported by both bioinformatic analysis and in vitro experiments. In terms of immune modulation, SIM2 may regulate the tumor microenvironment by altering immune cell infiltration, including Th17. Further experimental investigations should be warranted to substantiate the functional relevance of SIM2 and to unravel the underlying molecular mechanism in UCEC.
Data Sharing Statement
Data will be available from the corresponding author upon request.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This study was funded by Key Specialty Construction Project of Pudong Health and Family Planning Commission of Shanghai (Grant No.: PWZzk2022-14).
Disclosure
Yunfang Wei and Xianlei Zhao are co-first authors for this study. The authors have no relevant financial or non-financial interests to disclose for this work.
References
1. Crosbie EJ, Kitson SJ, McAlpine JN, Mukhopadhyay A, Powell ME, Singh N. Endometrial cancer. Lancet. 2022;399(10333):1412–1428. doi:10.1016/s0140-6736(22)00323-3
2. Gheorghe AS, Dumitrescu EA, Komporaly IA, Mihăilă RI, Lungulescu CV, Stănculeanu DL. New Targeted Therapies and Combinations of Treatments for Cervical, Endometrial, and Ovarian Cancers: a Year in Review. Curr Oncol. 2022;29(4):2835–2847. doi:10.3390/curroncol29040231
3. Tossetta G, Marzioni D. Targeting the NRF2/KEAP1 pathway in cervical and endometrial cancers. Eur J Pharmacol. 2023;941:175503. doi:10.1016/j.ejphar.2023.175503
4. Guo F, Zhang H, Jia Z, Cui M, Tian J. Chemoresistance and targeting of growth factors/cytokines signalling pathways: towards the development of effective therapeutic strategy for endometrial cancer. Am j Cancer Res. 2018;8(7):1317–1331.
5. Gu B, Shang X, Yan M, et al. Variations in incidence and mortality rates of endometrial cancer at the global, regional, and national levels, 1990-2019. Gynecologic Oncol. 2021;161(2):573–580. doi:10.1016/j.ygyno.2021.01.036
6. Tamaoki M, Komatsuzaki R, Komatsu M, et al. Multiple roles of single-minded 2 in esophageal squamous cell carcinoma and its clinical implications. Cancer Sci. 2018;109(4):1121–1134. doi:10.1111/cas.13531
7. Nakamura K, Komatsu M, Chiwaki F, et al. SIM2l attenuates resistance to hypoxia and tumor growth by transcriptional suppression of HIF1A in uterine cervical squamous cell carcinoma. Sci Rep. 2017;7(1):14574. doi:10.1038/s41598-017-15261-4
8. Kwak HI, Gustafson T, Metz RP, Laffin B, Schedin P, Porter WW. Inhibition of breast cancer growth and invasion by single-minded 2s. Carcinogenesis. 2007;28(2):259–266. doi:10.1093/carcin/bgl122
9. Aleman MJ, DeYoung MP, Tress M, Keating P, Perry GW, Narayanan R. Inhibition of Single Minded 2 gene expression mediates tumor-selective apoptosis and differentiation in human colon cancer cells. Proc Natl Acad Sci USA. 2005;102(36):12765–12770. doi:10.1073/pnas.0505484102
10. DeYoung MP, Tress M, Narayanan R. Down’s syndrome-associated Single Minded 2 gene as a pancreatic cancer drug therapy target. Cancer Lett. 2003;200(1):25–31. doi:10.1016/s0304-3835(03)00409-9
11. Halvorsen OJ, Rostad K, Øyan AM, et al. Increased expression of SIM2-s protein is a novel marker of aggressive prostate cancer. Clin Cancer Res. 2007;13(3):892–897. doi:10.1158/1078-0432.Ccr-06-1207
12. Launonen KM, Paakinaho V, Sigismondo G, et al. Chromatin-directed proteomics-identified network of endogenous androgen receptor in prostate cancer cells. Oncogene. 2021;40(27):4567–4579. doi:10.1038/s41388-021-01887-2
13. Uhlén M, Fagerberg L, Hallström BM, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. doi:10.1126/science.1260419
14. Oughtred R, Stark C, Breitkreutz BJ, et al. The BioGRID interaction database: 2019 update. Nucleic Acids Res. 2019;47(D1):D529–d541. doi:10.1093/nar/gky1079
15. Yoshihara K, Shahmoradgoli M, Martínez E, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. doi:10.1038/ncomms3612
16. Yang X, Li X, Cheng Y, et al. Comprehensive Analysis of the Glycolysis-Related Gene Prognostic Signature and Immune Infiltration in Endometrial Cancer. Front Cell Develop Biol. 2021;9:797826. doi:10.3389/fcell.2021.797826
17. Liew PL, Huang RL, Wu TI, et al. Combined genetic mutations and DNA-methylated genes as biomarkers for endometrial cancer detection from cervical scrapings. Clin epigenetics. 2019;11(1):170. doi:10.1186/s13148-019-0765-3
18. Patel MV, Shen Z, Rodriguez-Garcia M, Usherwood EJ, Tafe LJ, Wira CR. Endometrial Cancer Suppresses CD8+ T Cell-Mediated Cytotoxicity in Postmenopausal Women. Front Immunol. 2021;12:657326. doi:10.3389/fimmu.2021.657326
19. Golia D’Augè T, Cuccu I, Santangelo G, et al. Novel Insights into Molecular Mechanisms of Endometrial Diseases. Biomolecules. 2023;13(3):499. doi:10.3390/biom13030499
20. Cao W, Ma X, Fischer JV, Sun C, Kong B, Zhang Q. Immunotherapy in endometrial cancer: rationale, practice and perspectives. Biomarker Res. 2021;9(1):49. doi:10.1186/s40364-021-00301-z
21. Bogani G, Chiappa V, Lopez S, et al. Radiomics and Molecular Classification in Endometrial Cancer (The Rome Study): a Step Forward to a Simplified Precision Medicine. Healthcare. 2022;10(12):2464. doi:10.3390/healthcare10122464
22. Lefebvre TL, Ueno Y, Dohan A, et al. Development and Validation of Multiparametric MRI-based Radiomics Models for Preoperative Risk Stratification of Endometrial Cancer. Radiology. 2022;305(2):375–386. doi:10.1148/radiol.212873
23. Lu B, Asara JM, Sanda MG, Arredouani MS. The role of the transcription factor SIM2 in prostate cancer. PLoS One. 2011;6(12):e28837. doi:10.1371/journal.pone.0028837
24. Arredouani MS, Lu B, Bhasin M, et al. Identification of the transcription factor single-minded homologue 2 as a potential biomarker and immunotherapy target in prostate cancer. Clin Cancer Res. 2009;15(18):5794–5802. doi:10.1158/1078-0432.Ccr-09-0911
25. Scribner KC, Behbod F, Porter WW. Regulation of DCIS to invasive breast cancer progression by Singleminded-2s (SIM2s). Oncogene. 2013;32(21):2631–2639. doi:10.1038/onc.2012.286
26. DeYoung MP, Tress M, Narayanan R. Identification of Down’s syndrome critical locus gene SIM2-s as a drug therapy target for solid tumors. Proc Natl Acad Sci USA. 2003;100(8):4760–4765. doi:10.1073/pnas.0831000100
27. Bersten DC, Sullivan AE, Peet DJ, Whitelaw ML. bHLH-PAS proteins in cancer. Nat Rev Cancer. 2013;13(12):827–841. doi:10.1038/nrc3621
28. Berger MF, Mardis ER. The emerging clinical relevance of genomics in cancer medicine. Nat Rev Clin Oncol. 2018;15(6):353–365. doi:10.1038/s41571-018-0002-6
29. Tao MH, Freudenheim JL. DNA methylation in endometrial cancer. Epigenetics. 2010;5(6):491–498. doi:10.4161/epi.5.6.12431
30. Xu T, Ding H, Chen J, et al. Research Progress of DNA Methylation in Endometrial Cancer. Biomolecules. 2022;12(7):938. doi:10.3390/biom12070938
31. Salvesen HB, MacDonald N, Ryan A, et al. PTEN methylation is associated with advanced stage and microsatellite instability in endometrial carcinoma. Int j Cancer. 2001;91(1):22–26. doi:10.1002/1097-0215(20010101)91:1<22::aid-ijc1002>3.0.co;2-s
32. Arafa M, Kridelka F, Mathias V, et al. High frequency of RASSF1A and RARb2 gene promoter methylation in morphologically normal endometrium adjacent to endometrioid adenocarcinoma. Histopathology. 2008;53(5):525–532. doi:10.1111/j.1365-2559.2008.03147.x
33. Hsu YT, Gu F, Huang YW, et al. Promoter hypomethylation of EpCAM-regulated bone morphogenetic protein gene family in recurrent endometrial cancer. Clin Cancer Res. 2013;19(22):6272–6285. doi:10.1158/1078-0432.Ccr-13-1734
34. Tang J, Tian X, Min J, Hu M, Hong L. RPP40 is a prognostic biomarker and correlated with tumor microenvironment in uterine corpus endometrial carcinoma. Front Oncol. 2022;12:957472. doi:10.3389/fonc.2022.957472
35. Weijiao Y, Fuchun L, Mengjie C, et al. Immune infiltration and a ferroptosis-associated gene signature for predicting the prognosis of patients with endometrial cancer. Aging. 2021;13(12):16713–16732. doi:10.18632/aging.203190
36. Knochelmann HM, Dwyer CJ, Bailey SR, et al. When worlds collide: th17 and Treg cells in cancer and autoimmunity. Cell Mol Immunol. 2018;15(5):458–469. doi:10.1038/s41423-018-0004-4
37. Perez LG, Kempski J, McGee HM, et al. TGF-β signaling in Th17 cells promotes IL-22 production and colitis-associated colon cancer. Nat Commun. 2020;11(1):2608. doi:10.1038/s41467-020-16363-w
38. Kryczek I, Banerjee M, Cheng P, et al. Phenotype, distribution, generation, and functional and clinical relevance of Th17 cells in the human tumor environments. Blood. 2009;114(6):1141–1149. doi:10.1182/blood-2009-03-208249
39. Wang J, Li R, Cao Y, et al. Intratumoral CXCR5(+)CD8(+)T associates with favorable clinical outcomes and immunogenic contexture in gastric cancer. Nat Commun. 2021;12(1):3080. doi:10.1038/s41467-021-23356-w
40. Zou Q, Wang X, Ren D, et al. DNA methylation-based signature of CD8+ tumor-infiltrating lymphocytes enables evaluation of immune response and prognosis in colorectal cancer. J ImmunoTherapy Cancer. 2021;9(9):56.
41. Kondratiev S, Sabo E, Yakirevich E, Lavie O, Resnick MB. Intratumoral CD8+ T lymphocytes as a prognostic factor of survival in endometrial carcinoma. Clin Cancer Res. 2004;10(13):4450–4456. doi:10.1158/1078-0432.Ccr-0732-3
42. Palomero J, Panisello C, Lozano-Rabella M, et al. Biomarkers of tumor-reactive CD4(+) and CD8(+) TILs associate with improved prognosis in endometrial cancer. J ImmunoTherapy Cancer. 2022;10(12).
43. Janssen JBE, Medema JP, Gootjes EC, Tauriello DVF, Verheul HMW. Mutant RAS and the tumor microenvironment as dual therapeutic targets for advanced colorectal cancer. Cancer Treat Rev. 2022;109:102433. doi:10.1016/j.ctrv.2022.102433
44. van Gool IC, Eggink FA, Freeman-Mills L, et al. POLE Proofreading Mutations Elicit an Antitumor Immune Response in Endometrial Cancer. Clin Cancer Res. 2015;21(14):3347–3355. doi:10.1158/1078-0432.Ccr-15-0057
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.