Back to Journals » International Journal of Women's Health » Volume 16
Performance Evaluation of Noninvasive Prenatal Testing in Screening Chromosome Disorders: A Single-Center Observational Study of 15,304 Consecutive Cases in China
Authors Ye Q, Huang G, Hu Q, Man Q, Hao X, Liu L, Zhong Q, Jin Z
Received 19 December 2023
Accepted for publication 20 March 2024
Published 29 March 2024 Volume 2024:16 Pages 563—573
DOI https://doi.org/10.2147/IJWH.S455778
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Everett Magann
Qiang Ye,1 Guoping Huang,1 Qin Hu,1 Qin Man,2 Xiaoying Hao,3 Liangyan Liu,4 Qiang Zhong,1 Zhao Jin2
1Department of Clinical Laboratory, West China Second University Hospital, Southern Sichuan Women’s and Children’s Hospital, Zigong, Sichuan, 643000, People’s Republic of China; 2Department of Prenatal Diagnosis Center, West China Second University Hospital, Southern Sichuan Women’s and Children’s Hospital, Zigong, Sichuan, 643000, People’s Republic of China; 3Department of Ultrasound, West China Second University Hospital, Southern Sichuan Women’s and Children’s Hospital, Zigong, Sichuan, 643000, People’s Republic of China; 4Department of Obstetrics, West China Second University Hospital, Southern Sichuan Women’s and Children’s Hospital, Zigong, Sichuan, 643000, People’s Republic of China
Correspondence: Zhao Jin, Department of Prenatal Diagnosis Center, West China Second University Hospital, Southern Sichuan Women’s and Children’s Hospital, No. 49 Da Huang Tong Road, Da’an District, Zigong, Sichuan, 643000, People’s Republic of China, Tel +86-13990094008, Email [email protected]
Objective: This study was to evaluate the performance of noninvasive prenatal testing (NIPT) in detecting fetal chromosome disorders in pregnant women.
Methods: From October 1st, 2017, to December 31th, 2022, a total of 15,304 plasma cell free DNA-NIPT samples were collected for fetal chromosome disorders screening. The results of NIPT were validated by confirmatory invasive testing or clinical outcome follow-up. Further, NIPT performance between low-risk and high-risk groups, as well as singleton pregnancy and twin pregnancy groups was compared. Besides, analysis of 111 false-positive cases was performed.
Results: Totally, NIPT was performed on 15,086 eligible venous blood samples, of which 179 (1.19%) showed positive NIPT results and 68 were further validated to be true positive samples via confirmatory invasive testing or follow-up of clinical outcomes. For common chromosome aneuploidies, sex chromosome abnormalities (SCA) and other chromosomal aneuploidies, the detection sensitivities of NIPT were all 100%, the specificities were 99.87%, 99.70%, and 99.68% and the positive predictive values (PPVs) were 65.45%, 31.82%, and 10.91%, respectively. No statistically significant variance in detection performance was observed among 2987 high-risk and 12,099 low-risk subjects, as well as singleton and twin pregnancy subjects. The concentration of cell-free fetal DNA of 111 false-positive cases ranged from 5.5% to 33.7%, which was higher than the minimum requirement of NIPT.
Conclusion: With stringent protocol, NIPT shows high sensitivity and specificity for detecting fetal chromosome disorders in a large-scale clinical service, helping improving overall pregnancy management.
Keywords: noninvasive prenatal testing, fetal chromosome disorders, high risk, twin pregnancy, performance evaluation
Introduction
Fetal chromosome disorders are generally defined as syndromes caused by abnormal chromosome number or chromosome structure, leading to a complex and diverse range of symptoms such as intellectual disability, developmental delay, and malformations. Certain medications, treatments or rehabilitative methods can relieve the patient’s condition or suffering, but are not curative.1 Chromosome aneuploidy is the most common type of fetal chromosome disorders, among which trisomies (T) 21, 18, 13 are the most typical, and the incidence of these trisomies is relatively high, accounting for about 0.2% of all full-term pregnancies.2 Sex chromosome abnormalities (SCA), such as 47, XYY (Fernando syndrome), 47, XXX (Hyperemesis), 47, XXY (Klinefelter syndrome), 45, X (Turner syndrome), are a chromosomal disorder with an abnormal number of sex chromosomes, with an incidence of 1 in 200 to 1 in 400 live births and a prenatal diagnosis incidence of up to 1 in 435. Most of these patients are detected by karyotype analysis due to abnormal pubertal development, which may miss the best time to use the corresponding hormone treatment and lead to unremarkable treatment effect. In addition, with the development of treatment technology, other types of chromosomal disorders, such as other chromosome abnormalities and chromosome copy number variants (CNVs), are also attracting increasing attention.3
Prenatal screening is a vital tool to detect fetal chromosomal disorders and reduce the birth of fetuses with birth defects. Traditional screening and detection of fetal aneuploidy depends on biochemical detection and ultrasonic detection, and the detection rate is 50–95%,4 which is unstable and unsatisfactory, leading to many meaningless invasive tests. However, noninvasive prenatal testing (NIPT) has become widely available due to the discovery of free DNA from fetal cells in maternal plasma (cf-DNA) and the sustained development of next-generation sequencing technology (NGS). This method of screening provides several advantages, including being noninvasive and improved positive predictive value, accuracy, and safety.5–7 Currently, in some studies, NIPT has expanded beyond screening for common chromosome aneuploidies (T21, T18, T13) to include screening for SCA, and other chromosome abnormalities.6,8 Garshasbi et al, indicated that with a stringent protocol, NIPT showed excellent performance as screening test for the detection of fetal T21, T18, T13 and SCA in mixed-risk pregnancies in Iran.8 In Italy, a retrospective analysis of a large cohort of consecutive patients who had whole-genome sequencing-based NIPT for classic trisomies and SCA shows excellent detection rates and low false-positive rates.9 A retrospective analysis evaluated the performance of a new paired-end sequencing-based NIPT assay in 13,607 pregnancies from a single center in Germany, showing high sensitivities and specificities observed based on known clinical outcomes, a high overall PPV, and a low failure rate.10 However, there are limited studies in China which faced certain limitations such as small sample size, single chromosome disorders, and absent follow-up. Thus, the expanding application of NIPT has been controversial due to the potential for increased false-positive rates and the likelihood of uncertain findings with unclear clinical significance, which may contribute to patient distress and anxiety.11,12
Therefore, based on clinical data from 15,304 NIPT results in China, this study reported the detection performance of NIPT for the common chromosome aneuploidies, SCA and other chromosome abnormalities in a large-scale clinical service and compares the difference in detection performance of NIPT in high-risk and low-risk populations. Besides, this study analyzed the differences in detection performance of NIPT in singleton and twin pregnancy populations and further analyzed the characteristics of 111 false-positive cases, providing convincing evidence for the clinical application of NIPT.
Materials and Methods
Study Population and Design
Pregnant women who underwent conventional cf-DNA-based NIPT in West China Second University Hospital/Southern Sichuan Women’s and Children’s Hospital from October 1st, 2017, to December 31th, 2022, were recruited. The clinical data of the enrolled subjects were collected, including name, age, gestational age, ethnicity, pregnancy history, pregnancy mode, fetal ultrasound examination, NIPT results, clinical karyotype, pregnancy outcome, and neonatal physical examination report. Inclusion criteria: all enrolled subjects had singleton or twin pregnancy at 10 weeks of gestation or beyond and expressed a strong desire to undergo non-invasive prenatal testing (NIPT) screening. All subjects received professional and detailed genetic counseling before NIPT screening.
To encourage reporting of the false-positive (FP) and false-negative (FN) results, an insurance policy was provided for each participant as part of the test. Those who were found positive by NIPT were recommended to have confirmatory invasive testing. If refused, the pregnancy was monitored continuously to obtain pregnant outcome. In instances where the NIPT test yielded a negative result, yet subsequent laboratory tests, such as ultrasound, revealed chromosomal abnormalities, the policy offered a compensation of Chinese Yuan (CNY) 20,000. Should the NIPT test negated the presence of any chromosomal abnormalities, and subsequent ultrasound examinations corroborated this finding, yet significant chromosomal abnormality symptoms or positive genetic testing results were detected in the neonate or during follow-up visits, the policy would pay CNY 400,000 to each FN case.
When chromosome disorders were detected by any method, posttest genetic counseling was offered in all cases. This study was approved by the West China Second University Hospital/Southern Sichuan Women’s and Children’s Hospital Ethics Committee. Informed consent was taken from all individual participants.
NIPT
The NIPT procedures included the DNA extraction, library construction, whole genome sequencing, and bioinformatics analysis. Briefly speaking, 10 mL of blood samples was collected from each subjects and stored in tubes containing ethylenediaminetetraacetic acid. The cf-DNA was extracted from the samples using a Circulating Nucleic Acid Kit from Berry Genomics. Subsequently, the extracted plasma DNA was used as the input DNA to prepare the library for sequencing and quality control. Noninvasive DNA high-throughput sequencing technology was applied for sequencing, and the obtained sequences were compared with the human genome reference sequence map by bioinformatics analysis to detect common chromosome aneuploidies, SCA and other chromosome abnormalities. Fetal chromosome disorder risk was assessed by calculating the ratio Z-score. A high-risk result indicating sub-chromosomal deletions/duplications was indicated if the Z-score was |Z| >3, while a low-risk result was indicated if the Z-score was |Z| ≤3.
Confirmatory Invasive Testing
Prenatal invasive diagnosis that utilized both chromosomal karyotyping and CNV-seq was conducted. The amniocentesis was performed on positive subjects detected by NIPT at an average gestational age of 20 weeks. Chromosomal karyotyping was used to confirm whole chromosomal aneuploidies, and sub-chromosomal deletions and duplications were confirmed by CNV-seq. Samples were processed following established karyotyping protocols, including harvesting, colchicine treatment, digestion, fixation, preparation of spreads, and banding of chromosomes. We analyzed a minimum of 20 metaphases, and up to 50 metaphases were analyzed when mosaicism was detected. The sequencing reads were aligned to the human reference genome. CNV-seq was performed using NGS, which involved randomly interrupting genomic fragments with a restriction enzyme, end-filling, adding AMPs, linking to the primer, and ultimately sequencing and analyzing the fragments.
Comparison of NIPT Detection Performance Between Groups at Different Risk Levels
Subjects were divided into high-risk and low-risk groups according to the presence or absence of risk factors. Subjects in high-risk group could meet any of the following conditions: at least 35 years old; positive in conventional Down syndrome screening test; abnormal ultrasound indicators, family history of aneuploidy, and previous history of chromosomal abnormalities. Subjects without all these factors were defined as low-risk group. The detection performance of NIPT in the two groups was compared with the results of karyotype analysis or follow-up as the gold standard.
Comparison of NIPT Detection Performance Between Singleton Pregnancy and Twin Pregnancy Groups
A total of 15,086 subjects were divided into the singleton group and the twin group. The results of chromosome karyotype analysis or follow-up were used as the gold standard to compare the detection performance of NIPT between the two groups.
Collection of Postnatal Follow-Up Data
All the subjects were followed up by telephone and outpatient examination, and the registration form of subjects was reviewed to record the delivery status, pregnancy outcome, and neonatal health status (karyotype analysis and other cytogenetic results, physical examination reports).
Statistical Analysis
The data were analyzed using R software. For continuous data, normality was assessed using the Shapiro–Wilk method. When data met the normality assumption, they were presented as mean ± standard deviation. Those not conforming to a normal distribution were expressed as median (IQR). Categorical data were presented as percentages (%) and frequencies and compared using the chi-squared test or fisher exact test. A P value <0.05 was considered statistically significant. The detection performance (sensitivity, specificity, positive and negative predictive values) of the NIPT was determined using karyotyping results and clinical follow-up results as the gold standard, with 95% confidence intervals calculated. The true positive (TP) was defined as those positive NIPT results that were confirmed by prenatal invasive diagnosis or follow-up results. FP was defined as positive NIPT results that were shown to be normal euploidy karyotype or phenotype by the gold standard. The FN was defined as negative NIPT screening cases with an aneuploidy karyotype confirmed by prenatal invasive diagnosis or follow-up results. The true negative (TN) was defined as negative NIPT results confirmed by normal neonatal clinical physical examination results (barring SCA).
Results
Study Population Characteristics
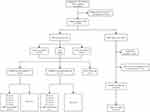
Peripheral blood samples from 15,304 subjects were included in the study. Out of the total samples, 140 (0.91%) were deemed unsuitable for further processing because of insufficient volume, contamination, or incorrect labeling. Seventy-eight samples (0.51%) required re-sampling because of failing quality control, or assay failure. Ultimately, 15,086 eligible samples (98.58%) were included (Figure 1).
The characteristics of the 15,086 subjects were shown in Table 1. The mean age of the pregnant women was 28.7 ± 4.60 years old, most of which (87.61%) were below 35 years old. Gestational ages at NIPT ranged from 11 to 36 weeks, with a mean of 18.15 weeks. The vast majority of the samples were collected in the second trimester (98.47%). In addition, there were 507 (3.36%) subjects of twin pregnancies and 14,579 (96.64%) subjects of singleton pregnancy among the 179 of the 15,086 samples (1.19%) had positive NIPT results, including 55 for common chromosome aneuploidies (T21, T18, and T13), 66 for SCA, and 58 for other chromosome abnormalities. Forty-nine underwent confirmatory invasive testing, which identified 28 true positive cases (including 7 cases of T21, 5 cases of T18, 1 case of T13, 5 cases of SCA, and 10 cases of other chromosomal abnormalities) and 21 false-positive cases. A total of 127 cases were verified through follow-up, resulting in the detection of 40 true positive cases (consisting of 20 cases of T21, 3 cases of T18, 16 cases of SCA, and 1 case of other chromosomal abnormalities) and 90 false-positive cases. To sum up, after confirmatory invasive testing and follow-up, 68 were true positive samples, 111 were false-positive samples, and 3 were lost to follow-up. Details could be available in Figure 1.
 |
Table 1 Demographic Characteristic of Pregnant Women Undergoing Noninvasive Prenatal Testing (NIPT) for Fetal Chromosome Disorders Between 2017 and 2022 |
As depicted in Table 2, among the individuals confirmed to be T21, 25 underwent selective abortion, 1 experienced neonatal death, and 1 delivered at term. Within the T18 true positive cases, 8 cases underwent selective abortion. One case was confirmed to be T13, and selective abortion was performed. Moreover, there were 5 types of SCA. Selective abortion was conducted in 18 cases, and term birth took place in 3 cases which were identified as XYY. Moreover, among the cases confirmed to be other chromosomal abnormalities, 4 cases underwent selective abortion, and 2 cases delivered at term. The remaining 14,907 cases with negative NIPT outcomes had follow-up results, and no false-negatives were discovered.
 |
Table 2 Pregnancy Outcomes of True Positive Cases Confirmed by Confirmatory Invasive Testing and Follow-Up Results |
Clinical Performance of NIPT in Testing Common Chromosome Aneuploidies
Among 15,086 subjects, 179 (1.19%) were NIPT positive. Fifty-five cases were found to be common chromosome aneuploidies. There were 33, 13 and 10 cases of T21, T18 and T13. The performance of NIPT in detecting common chromosome aneuploidies was shown in Table 3. In this subgroup, 27 true positive (TP) cases and 5 false-positive (FP) cases were detected for T21, while 8 TP cases and 5 FP cases were detected for T18 detection. For T13 detection, only 1 TP case and 9 FP cases were detected. Subsequently, for T21, T18, T13, the detection sensitivities of NIPT were all 100%, the specificities were 99.97%, 99.97%, and 99.94% and the positive predictive values (PPVs) were 84.38%, 61.53%, and 10.00%, respectively. In summary, the sensitivity, specificity and PPV of NIPT for the detection of common chromosome aneuploidies were 100%, 99.87% and 65.45%, respectively.
 |
Table 3 Performance of Noninvasive Prenatal Testing (NIPT) in Detecting Fetal Chromosome Disorders in Pregnancies with Outcome Data |
Clinical Performance of NIPT in Testing SCA and Other Chromosome Abnormalities
There were 66, 55 cases of SCA and other chromosome abnormalities detected by NIPT. Twenty-one TP cases and 45 FP cases were detected for SCA, while 6 TP case and 49 FP cases were detected for other chromosome abnormalities. Subsequently, for SCA and other chromosome abnormalities, the detection sensitivities of NIPT were all 100%, the specificities were 99.70%, and 99.68%, and the PPVs were 31.82%, and 10.91%, respectively.
Overall, the sensitivity, specificity and PPV of NIPT for fetal chromosome disorders were 100%, 99.27%, and 38.51%, respectively. Additionally, the theoretical PPV was calculated under two boundary conditions: all unconfirmed NIPT positive cases were assumed to be either true positive or false positive. Because there were no missing data in common chromosome aneuploidies, the theoretical PPV of T21, T18 and T13 were 84.38%, 61.54% and 10.00%, respectively. The theoretical PPV of SCA and other chromosome abnormalities were 31.82% and 10.34–15.52%, respectively. The overall theoretical PPV of all abnormalities was 36.31–37.99%.
Clinical Performance of NIPT in Testing Chromosomal Aneuploidies in Groups at Different Risk Levels
Based on the above criteria, 2987 of the 15,086 subjects were classified as high-risk and 12,099 as low-risk (Table 1). Particularly, a total of 3814 subjects underwent Down syndrome screening test and 1024 were found positive accounting for 34.28% in the high-risk group. Besides, 142 subjects (4.75%) had abnormal ultrasound indicators, and 1869 subjects (62.57%) were advanced maternal age. Besides, there was no family history of aneuploidy in the high-risk group.
The comparison of clinical performance of NIPT between the high-risk and low-risk groups was shown in Table 4. In the high-risk group, there were 46 positive cases detected by NIPT, including 23 TP cases, 22 FP cases, and 1 lost case, while there were 133 positive cases detected by NIPT, including 42 TP cases, 87 FP cases, and 4 lost cases in the low-risk group. Thus, for the high-risk group and in the low-risk group, the detection sensitivities of NIPT were all 100%, the specificities were 99.25% and 99.27%, and the PPVs were 51.11%, and 32.56%, respectively. No statistically significant variance in NIPT specificity was observed among 2987 high-risk and 2099 low-risk subjects (P = 0.905). However, the PPV of the high-risk group was significantly higher than that of the low-risk group (P = 0.026).
 |
Table 4 Performance of Noninvasive Prenatal Testing in Detection of Abnormalities in High-Risk Pregnancies and Low-Risk Pregnancies |
Clinical Performance of NIPT in the Detection of Chromosomal Aneuploidies in the Singleton Pregnancy and Twin Pregnancy Groups
As shown in Table 5, 507 subjects had twin pregnancies, and the remaining 14,579 subjects had singleton pregnancy. In the singleton pregnancy group, 174 cases had positive NIPT results, including 66 TP, 103 FP, and 5 lost follow-up. The sensitivity, specificity and PPV of NIPT were 100%, 99.29% and 39.05%, respectively. Likewise, in the twin pregnancy group, there were 5 positive cases detected by NIPT, including 1 TP case, 4 FP cases. The sensitivity, specificity and PPV of NIPT were 100%, 99.21% and 20.00%, respectively. No statistically significant variance in NIPT specificity and positive predictive value was observed among 2 groups.
 |
Table 5 Performance of Noninvasive Prenatal Testing in Detection of Abnormalities in Singleton Pregnancy and Twin Pregnancy Groups |
Analysis of false-positive Results
Totally, 111 false-positive results in subjects who were confirmed to be chromosome disorders after confirmatory invasive testing and clinical outcome follow-up were identified. The clinical genetic data of these 111 subjects were shown in Table 6. False-positive results were found in 21 cases by confirmatory invasive testing, and the remaining 86 cases were found by follow-up results. The median maternal age was 29.39 (18–41) years old. The median gestational age was 18.2 (12.6–28.1) weeks. There were 9 cases of common chromosome aneuploidies, 39 cases of SCA, and 42 cases of other chromosome abnormalities, including 4 twin pregnancy cases. The concentration of cell-free fetal DNA ranged from 5.5% to 33.7%, which was higher than the minimum requirement of NIPT.
 |
Table 6 Analysis of Biological Factors Causing 107 False-Positive Results |
Discussion
NIPT has been widely used to screen for T21, T18, and T13 in the past few years, yet relevant researches based on large clinical data is still deficient in China.13 Our study demonstrated that NIPT showed high sensitivity and specificity for detecting fetal chromosome disorders in a large-scale clinical service, which is consistent with previous results with small sample sizes.14 A recent study showed that NIPT had a sensitivity and specificity of more than 99.9% for detecting T21, T18, and T13, while the corresponding PPVs were 69.77%, 47.24%, and 22.36%.15 Likewise, in our study, the corresponding PPVs were 84.38%, 61.53% and 10.00%, respectively, and it also showed a downward trend, which may be due to the decline of the incidence of the three diseases. Thus, genetic counseling after NIPT should be strengthened for pregnant women with a high probability of abnormal test results to avoid misdiagnosis or misuse. Education and genetic counseling about prenatal screening for obstetricians and patients should be further strengthened.
Some extending disease-causing chromosome disorders could be detected by NIPT, such as SCA and other chromosome abnormalities. In this study, the sensitivity and specificity of NIPT in detecting SCA were 100% and 99.73%, which showed sound clinical performance. Additionally, recent studies reported PPVs for fetal SCA of over 50%,16 while the PPV of SCA was 38.46% in our study, which was lower than 50%, probably due to the pregnant woman’s clinical profile and the characteristics of the testing laboratory. Moreover, cf-DNA screening allowed not only the detection of common chromosome abnormalities but also enabled the identification of other chromosome abnormalities. In this study, the sensitivity and specificity of NIPT detection for other chromosome abnormalities were 100%, and 99.68%, respectively. Such additional data may be helpful in improving pregnancy management. However, the PPV was only 11.11%, which may be attributing to its low incidence and needs more samples to verify. Overall, the need to perform invasive testing in cases with SCA and other chromosome abnormalities should be discussed between geneticists, obstetricians and parents, eventually allowing the development of rules for the best clinical follow-up actions to be taken.17
Particularly, there is growing attention in the detection performance of NIPT in different populations. Previous study has found that the specificity of NIPT in low-risk pregnancies is better than that in high-risk pregnancies, but this difference does not seem to be clinically meaningful.18 Similarly, in our study, the sensitivity and specificity of detection in high-risk and low-risk groups are equivalent, which proves that NIPT is reliable as a routine screening test in various kinds of pregnant women. Besides, in this study, the positive predictive value PPV was significantly higher in the high-risk group than in the low-risk group, which is consistent with earlier studies and may be due to the reduced PPV in the low-risk group due to lower disease incidence.19 Studies have shown that twin pregnancies are more likely to have fetal structural abnormalities than singleton pregnancies and that twin pregnancies have specific complications during pregnancy.20 In this study, a total of 504 twin pregnancies were included, and the results showed that the sensitivity of NIPT in the detection of both singleton and twin pregnancies groups was 100%. Notably, the specificity in the latter group was slightly lower, and the PPV was also lower than that in the singleton pregnancy group, which may be because twin pregnancy is more prone to placenta mosaic and leads to false positives. Moreover, NIPT can safely be offered to women who are pregnant with twin pregnancies since in addition to the risk of an abnormal result, the risk of invasive test procedures is higher than the singleton pregnancy women.
However, NIPT is only a screening method after all, and there may be false positives in the test results. Confounding factors such as confined placental mosaicism, maternal chromosomal anomalies, and maternal background of CNV and CPM may be attributed to the false positives.20,21 Therefore, when interpreting NIPT results, it is vital to take into account these factors. Pregnant subjects should be provided with professional post-test genetic counseling to ensure a thorough understanding of the results.22 In this study, the concentration of cell-free fetal DNA of all subjects ranged from 5.5 to 33.7%, which was higher than the minimum requirement of NIPT. Actually, placental mosaicism is associated with intrauterine growth restriction (IUGR), small-for-gestational-age infants, and unfavorable pregnancy outcomes.23,24 As a result, when counseling patients with positive NIPT results for uncommon aneuploidies, the possibility of fetal mosaicism, CPM, and uniparental disomy (UPD) should be discussed. In such cases, chromosomal karyotyping and CNV-seq should be performed, and serial ultrasound examinations should be conducted to monitor fetal growth.25
There are also several limitations in this study. First, a considerable number of NIPT positive subjects were unwilling to accept invasive testing. Besides, not all subjects underwent Down syndrome screening test. In addition, only one hospital’s patient data was included in this study, and selection bias, data loss due to pregnancy termination and loss to follow-up cannot be avoided. Larger-scale, multicenter studies in real-world settings are needed to provide a more comprehensive exploration for NIPT performance.
Conclusion
Currently, this study provides a true reflection of what NIPT could achieve in large scale of practice. With strict protocol and quality management, our study showed a high sensitivity and specificity of NIPT in detecting fetal chromosome disorders, especially the common chromosome aneuploidies, showing great potential for clinical application. However, caution should be paid when consulting NIPT results for twin pregnancies, positive SCA, or other chromosomal abnormalities and require other invasive tests to assist in interpreting the results.
Data Sharing Statement
All data generated or analyzed were included in this published article.
Ethics Approval and Consent to Participate
This study was approved by the West China Second University Hospital/Southern Sichuan Women’s and Children’s Hospital Ethics Committee. Informed consent was taken from all individual participants.
Consent for Publication
All authors approved the final manuscript and consented to publish this manuscript.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This study was financially supported by the Scientific and Technological Innovation Project in Maternal and Child Medicine of Sichuan Province (22FXYB05).
Disclosure
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Shorey S, Lalor J, Pereira TL, Jarašiūnaitė-Fedosejeva G, Downe S. Decision-making and future pregnancies after a positive fetal anomaly screen: a scoping review. J Clin Nurs. 2023;32(17–18):5534–5549.
2. Bianchi DW, Oepkes D, Ghidini A. Current controversies in prenatal diagnosis 1: should noninvasive DNA testing be the standard screening test for down syndrome in all pregnant women? Prenat Diagn. 2014;34(1):6–11. doi:10.1002/pd.4229
3. Xiang P, Liu L, Hu X, Zhou Y. CNV-seq 在高危孕妇产前诊断中的应用价值 [Application value of CNV-seq for the prenatal diagnosis of women with high-risk pregnancies]. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2023;40(1):17–20. Chinese. doi:10.3760/cma.j.cn511374-20211209-00976
4. Dan S, Wang W, Ren J, et al. Clinical application of massively parallel sequencing-based prenatal noninvasive fetal trisomy test for trisomies 21 and 18 in 11,105 pregnancies with mixed risk factors. Prenat Diagn. 2012;32(13):1225–1232. doi:10.1002/pd.4002
5. Cernat A, De Freitas C, Majid U, Trivedi F, Higgins C, Vanstone M. Facilitating informed choice about non-invasive prenatal testing (NIPT): a systematic review and qualitative meta-synthesis of women’s experiences. BMC Pregnancy Childbirth. 2019;19(1):27. doi:10.1186/s12884-018-2168-4
6. Zhang Y, Xu H, Zhang W, Liu K. Non-invasive prenatal testing for the detection of trisomy 13, 18, and 21 and sex chromosome aneuploidies in 68,763 cases. Front Genetics. 2022;13:864076. doi:10.3389/fgene.2022.864076
7. Saidel ML, Ananth U, Rose D, Farrell C. Non-invasive prenatal testing with rolling circle amplification: real-world clinical experience in a non-molecular laboratory. J Clin Lab Analysis. 2023;37(6):e24870. doi:10.1002/jcla.24870
8. Garshasbi M, Wang Y, Hantoosh Zadeh S, Giti S, Piri S, Reza Hekmat M. Clinical application of cell-free DNA sequencing-based noninvasive prenatal testing for trisomies 21, 18, 13 and sex chromosome aneuploidy in a mixed-risk population in Iran. Fetal Diagn Ther. 2020;47(3):220–227. doi:10.1159/000501014
9. La Verde M, De Falco L, Torella A. Performance of cell-free DNA sequencing-based non-invasive prenatal testing: experience on 36,456 singleton and multiple pregnancies. BMC Med Genomics. 2021;14(1):93. doi:10.1186/s12920-021-00941-y
10. Borth H, Teubert A, Glaubitz R, et al. Analysis of cell-free DNA in a consecutive series of 13,607 routine cases for the detection of fetal chromosomal aneuploidies in a single center in Germany. Arch Gynecol Obstetrics. 2021;303(6):1407–1414. doi:10.1007/s00404-020-05856-0
11. Christiaens L, Chitty LS, Langlois S. Current controversies in prenatal diagnosis: expanded NIPT that includes conditions other than trisomies 13, 18, and 21 should be offered. Prenat Diagn. 2021;41(10):1316–1323. doi:10.1002/pd.5943
12. Jani JC, Gil MM, Benachi A. Genome-wide cfDNA testing of maternal blood. Ultrasound Obstet Gynecol. 2020;55(1):13–14. doi:10.1002/uog.21945
13. Gregg AR, Skotko BG, Benkendorf JL, et al. Noninvasive prenatal screening for fetal aneuploidy, 2016 update: a position statement of the American college of medical genetics and genomics. Genet Med. 2016;18(10):1056–1065. doi:10.1038/gim.2016.97
14. Gu Z, Du M, Xu T, Jin C, Tang M. Study on the clinical value of noninvasive prenatal testing in screening the chromosomal abnormalities of the fetus in the elderly pregnant women. Comput Math Methods Med. 2022;2022:2977128. doi:10.1155/2022/2977128
15. Xiang L, Zhu J, Deng K. Non-invasive prenatal testing for the detection of trisomies 21, 18, and 13 in pregnant women with various clinical indications: a multicenter observational study of 1,854,148 women in China. Prenat Diagn. 2023;43(8):1036–1043. doi:10.1002/pd.6312
16. Mennuti MT, Chandrasekaran S, Khalek N, Dugoff L. Cell-free DNA screening and sex chromosome aneuploidies. Prenat Diagn. 2015;35(10):980–985. doi:10.1002/pd.4639
17. Fiorentino F, Bono S, Pizzuti F, et al. The clinical utility of genome-wide non invasive prenatal screening. Prenat Diagn. 2017;37(6):593–601. doi:10.1002/pd.5053
18. Miao Z-Y, Liu X, Shi T-K, Ge J-M, Xu Y. 早、中孕期整合筛查唐氏综合征 [First trimester and second-trimester integrated screening for Down’s syndrome]. Zhonghua Yi Xue Za Zhi. 2011;91(3):185–188. Chinese.
19. Liu S, Chang Q, Yang F, et al. Non-invasive prenatal test findings in 41,819 pregnant women: results from a clinical laboratory in southern China. Arch Gynecol Obstetrics. 2023;308(3):787–795. doi:10.1007/s00404-022-06908-3
20. Konishi A, Samura O, Muromoto J. Prevalence of common aneuploidy in twin pregnancies. J Hum Genet. 2022;67(5):261–265. doi:10.1038/s10038-021-01001-0
21. Yang L, Tan WC. Prenatal screening in the era of non-invasive prenatal testing: a nationwide cross-sectional survey of obstetrician knowledge, attitudes and clinical practice. BMC Pregnancy Childbirth. 2020;20(1):579. doi:10.1186/s12884-020-03279-y
22. Montgomery S, Thayer ZM. The influence of experiential knowledge and societal perceptions on decision-making regarding non-invasive prenatal testing (NIPT). BMC Pregnancy Childbirth. 2020;20(1):630. doi:10.1186/s12884-020-03203-4
23. Robinson WP, Peñaherrera MS, Jiang R, et al. Assessing the role of placental trisomy in preeclampsia and intrauterine growth restriction. Prenat Diagn. 2010;30(1):1–8. doi:10.1002/pd.2409
24. Wilkins-Haug L, Quade B, Morton CC. Confined placental mosaicism as a risk factor among newborns with fetal growth restriction. Prenat Diagn. 2006;26(5):428–432. doi:10.1002/pd.1430
25. Lau TK, Cheung SW, Lo PS, et al. Non-invasive prenatal testing for fetal chromosomal abnormalities by low-coverage whole-genome sequencing of maternal plasma DNA: review of 1982 consecutive cases in a single center. Ultrasound Obstet Gynecol. 2014;43(3):254–264. doi:10.1002/uog.13277
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.