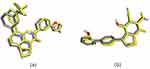Back to Journals » Journal of Experimental Pharmacology » Volume 15
Molecular Docking Study of Phytosterols in Lygodium microphyllum Towards SIRT1 and AMPK, the in vitro Brine Shrimp Toxicity Test, and the Phenols and Sterols Levels in the Extract
Authors Anggreini P , Kuncoro H , Sumiwi SA, Levita J
Received 30 September 2023
Accepted for publication 19 December 2023
Published 22 December 2023 Volume 2023:15 Pages 513—527
DOI https://doi.org/10.2147/JEP.S438435
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Tuo Deng
Putri Anggreini,1,2,* Hadi Kuncoro,2,* Sri Adi Sumiwi,3,* Jutti Levita3,*
1Faculty of Pharmacy, Padjadjaran University, Sumedang, 46363, Indonesia; 2Faculty of Pharmacy, Mulawarman University, Samarinda, 75119, Indonesia; 3Department of Pharmacology and Clinical Pharmacy, Faculty of Pharmacy, Padjadjaran University, Sumedang, 46363, Indonesia
*These authors contributed equally to this work
Correspondence: Jutti Levita, Department of Pharmacology and Clinical Pharmacy, Faculty of Pharmacy, Padjadjaran University, Sumedang, West Java, 46363, Indonesia, Tel +6222-84288888 Ext 3510, Email [email protected]
Background: Lygodium microphyllum is a fern plant with various pharmacological activities, and phytosterols were reported contained in the n-hexane and ethyl acetate extract of this plant. Phytosterols are known to inhibit steatosis, oxidative stress, and inflammation. Sirtuin 1 (SIRT1) and adenosine monophosphate-activated protein kinase (AMPK) are the key proteins that control lipogenesis. However, information about L. microphyllum on SIRT1 and AMPK is still lacking.
Purpose: This study aims to investigate the binding mode of phytosterols in L. microphyllum extract towards AMPK and SIRT1, and the toxicity of the extract against brine shrimp (Artemia salina) larvae, and to determine the phenols and sterols levels in the extract.
Methods: The molecular docking was performed towards SIRT1 and AMPK using AutoDock v4.2.6, the toxicity of the extract was assayed against brine shrimp (Artemia salina) larvae, and the phytosterols were analyzed by employing a thin layer chromatography densitometry, and the total phenols were by spectrophotometry.
Results: The molecular docking study revealed that β-sitosterol and stigmasterol could occupy the active allosteric-binding site of SIRT1 and AMPK by binding to important residues similar to the protein’s activators. The cold extraction of the plant yields 15.86% w/w. Phytochemical screening revealed the presence of phenols, steroids, flavonoids, alkaloids, and saponins. The total phenols are equivalent to 126 mg gallic acid (GAE)/g dry extract, the total sterols are 954.04 μg/g, and the β-sitosterol level is 283.55 μg/g. The LC50 value of the extract towards A. salina larvae is 203.704 ppm.
Conclusion: Lygodium microphyllum extract may have the potential to be further explored for its pharmacology activities, particularly in the discovery of plant-based anti-dyslipidemic drug candidates. However, further studies are needed to confirm their roles in alleviating lipid disorders.
Keywords: anti-dyslipidemia, cholesterol, inflammation, lipogenesis, phytosterols
Introduction
An epidemiology study reported that lipid disorders such as non-alcoholic liver disease (NAFLD) had reached 13.5% (Africa) to 31.8% (Middle East) of cases. The elevating prevalence of this disease is correlated with the global obesity epidemic and the manifestation of metabolic disorders.1 Lipid disorders occur due to unhealthy diet patterns such as high fat or high sugar intake and a sedentary lifestyle. People with fatty liver disease are prone to suffer vascular risk and cardiovascular disease progression.2 The Food and Drug Administration (FDA)-approved drug for NAFLD is limited.3 Therefore, finding new drugs for NAFLD therapy is challenging.
Several proteins are responsible for the development of NAFLD, specifically in the lipogenesis mechanism, such as Sirtuin 1 (SIRT1) and adenosine monophosphate-activated protein kinase (AMPK).4 SIRT1 is a histone deacetylase protein that is involved in several biological processes including the antioxidant system, inflammation, cell death program, and lipid-glucose metabolism.5,6 Recent studies confirm that the activation of SIRT1 may interfere with the lipogenesis process, resulting in the lowering of lipid levels.6–8 AMPK, as a regulatory protein, regulates metabolism in the cell, thus it plays a major role in preventing the occurrence of excess fat production.9,10 A previous study reported that the increased expression of AMPK is in line with the decrease in several adipogenic proteins such as fatty acid synthase (FAS) and stearoyl-CoA desaturase 1 (SCD1).11 Furthermore, the administration of a compound that activates AMPK, such as PXL770 (a thienopyridine drug for oral use that is undergoing clinical study), resulted in a reduction of several parameters of fat and de novo lipogenesis percentage in NAFLD patients.12
Numerous studies have reported that phytosterols may contribute to arresting the onset and progression of NAFLD although their mechanisms are not known.13 Lygodium microphyllum is a fern plant that has been studied for several pharmacological activities.14–16 Phytosterols, eg, stigma-5(6)-en-3β-ol and stigmast-4-en-3-one, have been isolated from the n-hexane and ethyl acetate extract of L. microphyllum.16,17 However, information about the effects of phytosterols on AMPK and SIRT1 is still lacking. Considering this, our study aimed to investigate the binding mode of these phytosterols towards SIRT1 and AMPK by in silico approaches and to assay the toxicity of L. microphyllum extract using brine shrimp (Artemia salina) larvae, total sterols, and total phenols, to provide a thorough preliminary data of L. microphyllum.
Materials and Methods
Preparation and Phytochemical Screening of the Extract
Briefly, L. microphyllum herbs (Figure 1) were collected from a rainforest in East Kalimantan, Indonesia, in July 2022. The herbs were determined by Dr. Atiek Retnowati and were deposited in Herbarium Bogoriense, the Research Center for Biology, the Indonesian Institute of Sciences, Indonesia, and were confirmed as Lygodium microphyllum (Lygodiaceae) (voucher specimen number 750IPH101). The characteristics of the plant samples matched those described in The IUCN Red List of Threatened Species (https://www.iucnredlist.org/species/194153/8883960).
 |
Figure 1 The fern plant Lygodium microphyllum (Lygodiaceae), collected from a rainforest in Samarinda, East Kalimantan, Indonesia. |
The herbs (approximately 3.6 kg of weight) were extracted with ethanol 70% as the solvent with a ratio of 1:3 w/v for 3×24 h at room temperature. Ethanol was used as the extraction solvent due to its universal characteristics in dissolving almost all plant secondary metabolites. The extracts were filtered using a Whatman paper and the solvent was rotary-evaporated (BUCHI) in a vacuum at 50 °C until a viscous extract (571 g) of L. microphyllum (abbreviated as ELM) was yielded (15.86% w/w).
Chemicals
The chemicals used in this study were ethanol analytical grade (Merck CAS 64-17-5100983), chloroform (Merck CAS 67-66-3102445), acetic acid (Merck CAS 64-19-7100056), Mayer’s reagent (Sigma-Aldrich), Dragendorff’s reagent (Sigma-Aldrich), sulfuric acid (Merck CAS 7664-93-9100731), sodium hydroxide (Merck), sodium carbonate (Merck), magnesium turnings (Sigma-Aldrich), Folin-Ciocalteu’s reagent (Merck), gallic acid (Merck), dimethyl sulfoxide (DMSO), beta-sitosterol (Sigma-Aldrich), toluene (Merck), ethyl acetate (Merck), Lieberman Burchard’s reagent (Merck).
Determination of the Total Phenolic Content
The determination of the total phenolic content (TPC) was carried out by following a previous method.18 Briefly, a solution of the extract of L. microphyllum (ELM; 200 ppm) was added with Folin-Ciocalteu reagent and then incubated within 1 minute. The solution was added with Na2CO3 2% and then incubated within 30 minutes. The solution was further analyzed using a UV–visible spectrophotometer (Dynamica HALO DB 20S) at 510 nm. Gallic acid was used as a standard with concentration series of 25 ppm, 50 ppm, 100 ppm, and 200 ppm. The result is expressed in gallic acid equivalents (GAE)/g dry extract weight.
Determination of Total Sterols
The determination of total sterols was carried out by following a previous method.19 Approximately 50 mg of ELM was placed in a microtube, added with 1 mL of ethanol until dissolved, vortexed for 30 seconds, sonicated for 60 minutes, and kept for 24 hours at 26–28 °C. 1 µL of the supernatant was taken using a micropipette after centrifugation and was put on silica gel 60 F254 together with the beta-sitosterol standard (Sigma-Aldrich) in various concentrations (0.6 µg, 1.2 µg, 2.4 µg, 3.6 µg, 4.8 µg, and 6.0 µg). The plate was inserted into a chamber saturated with a mixture of toluene and ethyl acetate (80:20) as the mobile phase, eluted to the finish line, and dried. The plate was then sprayed with the Liebermann-Burchard reagent (Merck) containing acetic anhydride and sulfuric acid, which is specific to detect the presence of sterol compounds. The sprayed plate with separated bands was heated at 110 °C for 2 minutes, and the bands were captured and further analyzed with a densitometer. The data of the peak such as height, area, and Rf values were recorded, and the concentration of total steroids and beta-sitosterol in ELM was calculated.
Brine Shrimp Toxicity Test
The premium-grade brine shrimp (Artemia salina) eggs were obtained from an ornamental fish shop in Samarinda, East Kalimantan, Indonesia. The brine shrimp larvae were obtained by placing the eggs in a seawater medium (pH > 8) in a round glass container. An aerator was placed inside the container to provide sufficient oxygen levels and a lamp to maintain the appropriate temperature for hatching (26–28 °C). In 48 hours, the larvae were ready to be used for the study.
The toxicity was evaluated following a previous method.20 ELM was dissolved in ethanol at increasing concentrations (100 ppm, 150 ppm, 200 ppm, 250 ppm, and 300 ppm) and added with ten brine shrimp larvae. The ELM and the larvae were incubated for 24 hours at room temperature. After 24 hours after exposure, the number of survival larvae (LC50) was calculated by determining the probit value of death percentage in each group of the test, using a linear regression equation obtained from the graph (log concentration as the x-axis plotted against probit value as the y-axis).
Molecular Docking Simulation
Hardware and Software
The hardware used was an HP Pavilion laptop with Intel CORE i7 processor Gen 8 RAM 16GB SSD 512GB. The software used were the AutoDock-v4.2.6 for Windows (https://autodock.scripps.edu/download-autodock4/), the AutoDockTools 1.5.6, a PerkinElmer ChemDraw-Pro-v15.00, and a Discovery Studio Visualizer 4.0.
Protein and Ligand Preparation
The protein 3D structures of human SIRT1 (PDB ID: 4ZZI, resolution 2.73 Å in the complex with 4TQ, https://www.rcsb.org/structure/4ZZI) and human AMPK (PDB ID: 4CFF, resolution 3.92 Å in the complex with A-769662, https://www.rcsb.org/structure/4cff) were acquired from the Protein Data Bank (https://www.rcsb.org/).21,22 The native ligands, 4TQ and A-769662, were detached from the proteins, while other ligands, substrates, ions, waters, and hetero atoms, were abolished using the Discovery Studio program. The 2D structures of beta-sitosterol and stigmasterol were prepared using the ChemDraw Pro-v15, then were re-modeled to the 3D structures, optimized by applying the Merck molecular force field (MMFF94) and reserved to a.pdb format. The proteins and ligands were constructed and changed to the PDB-partial charge (q)-atom type (t) format (.pdbqt) using AutoDockTools.
Validation of the Molecular Docking Simulation
Validation was performed by re-docking the 4TQ and A-769662 into their previous sites and continued by superimposing the re-docked conformers with the original poses. The root mean square deviation (RMSD) of the structures was calculated.23,24
Molecular Docking
Following the protocols, beta-sitosterol and stigmasterol were docked at the binding site of SIRT1 and AMPK with 100x iterations. The grid box dimension of 40 x 40×40 Å was used on the SIRT1 protein, centered at the active site: X = −0.827, Y = 45.618, Z = −0.546, at a distance of 0.375 Å. The grid box dimension of 50 x 50×50 Å was used on the AMPK protein, centered at the active site: X = −22.682, Y = −9.953, Z = 207.822, at a distance of 0.375 Å. Other parameters were fixed at an automatic preselected setting. The docking results were analyzed using AutoDockTools and visualized using the Discovery Studio Visualizer 4.0. Parameters observed were (1) the free binding energy (ΔG) which represents the affinity between the ligand and the macromolecule and (2) the binding mode with important amino acid residues, in terms of hydrogen bond and hydrophobic interaction.
Results
Phytochemical Screening and Total Phenols
The phytochemical screening of ELM confirmed the presence of steroids, polyphenols, flavonoids, alkaloids, and saponins. The total phenols in ELM calculated using a gallic acid standard curve (linear regression equation 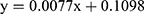 ; R2 = 0.9951) is 126 mg GAE/g dry extract.
; R2 = 0.9951) is 126 mg GAE/g dry extract.
Total Sterols and Beta-Sitosterol Levels
The thin-layer chromatography bands revealed the presence of four phytosterols in the L. microphyllum herbs extract (replicated three times and labeled as ELM1, ELM2, and ELM3) (Figure 2) with an Rf value of 0.20, 0.47, 0.50, and 0.75. Compared to the standard β-sitosterol that is eluted at Rf 0.50 (replicated four times and labeled as BS1, BS2, BS3, and BS4), ELM is confirmed to contain β-sitosterol in a considerable amount, whereas the band eluted at Rf 0.47 is predicted belong to stigmasterol due to their close similarity in their chemical structure (Figure 3). The TLC-densitometry chromatograms of sterols in ELM and standard β-sitosterol are depicted in Figure 4, which proves that ELM contains four different sterols (the under-curve areas of 5233.05 AU, 3934.1 AU, 1005.95 AU, and 1018.15 AU, respectively). β-sitosterol was identified in ELM with the Rf value of 0.50 and 3934.1 AU. The area of the sterol compound was further calculated using a formula from the β-sitosterol standard curve (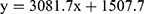 ; R2 = 0.9948) and resulted in total sterols of 954.04 µg/g and the amount of β-sitosterol is 283.55 µg/g.
; R2 = 0.9948) and resulted in total sterols of 954.04 µg/g and the amount of β-sitosterol is 283.55 µg/g.
 |
Figure 4 TLC-densitometry chromatograms of β-sitosterol standard with increasing concentrations (a–f), beta-sitosterol in ELM (BS), and other steroids in ELM (S1-S3). |
Brine Shrimp Toxicity Test
The toxicity of ELM screened using the brine shrimp toxicity test is tabulated in Table 1. The highest percentage of mortality (5.6%) occurred in ELM 300 ppm while the lowest was 0.2% (DMSO). The LC50 of ELM (determined by employing the logarithmic concentration of ELM vs probit value of % mortality; 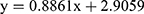 ; R2 = 0.9504) is 203.704 ppm.
; R2 = 0.9504) is 203.704 ppm.
 |
Table 1 Brine Shrimp Toxicity Test of ELM |
Molecular Docking Simulation
Considering that phytosterols are contained in the extract, we performed a molecular docking simulation of β-sitosterol and stigmasterol with SIRT1 (PDB ID 4ZZI) and AMPK (PDB ID 4CFF), which was started with the validation step. The validity of the molecular docking program is proven by the root mean square deviation (RMSD) value <2.0 Å (depicted in Figure 5). The re-docking of 4TQ and A-769662 to SIRT1 and AMPK, respectively, reveals the type of interaction and bond distance with important amino acid residues as presented in Table 2.
 |
Table 2 The Affinity in Terms of the Docking Score and the Interaction of the Ligands with Important Residues of SIRT1 and AMPK Compared to the Allosteric Activators |
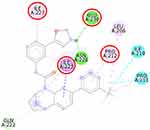
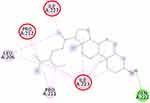
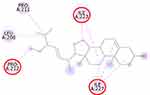
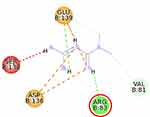
The molecular docking simulation of β-sitosterol and stigmasterol with SIRT1 and AMPK is tabulated in Table 2. The allosteric ligand of SIRT1, namely 4TQ (Figure 6), builds two hydrogen bonds with Glu230 and Asn226, and seven hydrophobic interactions with essential amino acid residues in the allosteric regions of SIRT1; thus, it shows the lowest binding energy (−7.27 kcal/mol). The hydrogen bond interaction with Glu230 and Gln 222 residues is demonstrated by resveratrol (Figure 7) but with a higher binding energy (−4.05 kcal/mol). The 2D molecular interactions of β-sitosterol (−6.11 kcal/mol) and stigmasterol (−6.45 kcal/mol) with SIRT1 are depicted in Figures 8 and 9, respectively. The bond distance between ligands and amino acid residues is summarized in Table 2.
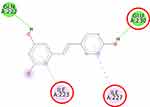
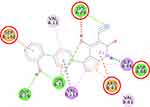
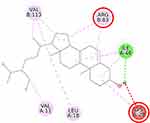
Furthermore, the 2D molecular interaction of the native ligand of AMPK, namely A-769662 (Figure 10), also possesses the lowest binding energy (−9.36 kcal/mol), whereas metformin (Figure 11) shows the highest binding energy (−3.50 kcal/mol). The 2D molecular interaction β-sitosterol (−8.15 kcal/mol) and stigmasterol (−9.04 kcal/mol) with AMPK are depicted in Figures 12 and 13, respectively. Interestingly, both β-sitosterol and stigmasterol show a stronger affinity towards AMPK compared to that of metformin.
All things considered, our molecular docking study showed that both β-sitosterol and stigmasterol could interact with important residues of SIRT1 and AMPK. These phytosterols show stronger affinity, in terms of lower binding energy, compared to that of resveratrol and metformin.
Discussion
L. microphyllum is a plant that has a lot of potential to become a medicinal plant. Our present study confirmed that the ethanol extract of L. microphyllum (ELM) contains important secondary metabolites such as steroids, phenols, flavonoids, alkaloids, and saponins. In previous studies, L. microphyllum extracted with methanol also showed similar secondary metabolites such as steroids, flavonoids, alkaloids, and saponins.25
Secondary metabolites, such as phytosterols, have been approved by the FDA as lipid-lowering agents.26 The phenol and flavonoid groups are known for their activity as antioxidants. Phenol compounds have been widely reported to exhibit various pharmacological activities such as antioxidant, anti-inflammatory, anti-neurological disorder, anti-diabetic, anti-cancer, and many more.27
In the present study, the total phenols in ELM are 126 mg GAE/g of dry extract which is comparable to a previous study on L. lanceolatum (101.68 mg GAE/g).28 The high phenolic content of ELM indicates that L. microphyllum may be beneficial for combating several diseases related to oxidative stress, due to the ability of phenolic compounds to donate a proton to scavenge free radicals or their mechanism as antioxidants. In agreement with a previous study, the doses of 200 and 400 mg/kg of the methanol extract of L. microphyllum had improved disease related to oxidative stress such as inflammation, pain disorder, and diarrheal.14
Our study revealed that the sterols measured using the TLC-densitometry technique were 954.04 µg/g. This study is the first that reported the amount of beta-sitosterol in the ethanol extract of L. microphyllum (ELM), which is 283.55 µg/g. A fairly high sterol content in plants might indicate the potential of the plant against lipid disorders. Plant sterols have been reported to have good benefits in improving lipid metabolism.29,30 Phytosterols administered as add-on therapy at a dose of 2 g per day have demonstrated their capacity to lower LDL-cholesterol in patients with dyslipidemia, and hence, are suggested in high-risk patients who have failed by statins.3 The principal mechanism of phytosterols in lowering LDL-cholesterol levels is by (1) reducing the intestinal absorption of cholesterol similar to ezetimibe,31 (2) decreasing the rate of cholesterol esterification in the enterocyte similar to statins, and (3) elevating the elimination of cholesterol via transintestinal excretion similar to the fibrates.32
Our study demonstrates that ELM (with an LC50 of 203.704 ppm) is categorized as moderate toxicity (LC50 > 30 to 1000 ppm).33 However, toxicity tests with advanced methods must be carried out to elucidate further information.
A study reported that β-sitosterol has been successfully isolated from L. microphyllum as the main phytosterol in this fern, apart from that there is also stigmasterol.17 Up to this point, no molecular docking simulation on phytosterols SIRT1 and AMPK was reported, thus implying the present study is the first. The molecular docking simulation showed that both β-sitosterol and stigmasterol could interact with important residues of SIRT1 and AMPK. These phytosterols show stronger affinity, in terms of lower binding energy, compared to that of resveratrol and metformin. The binding energy determines how strong and stable the interaction between the ligand and the protein target is. The lower the value, the smaller the amount of energy required to form a complex.34 Interestingly, the molecular docking studies of phytosterols contained in many plants, eg, Polygonum hydropiper, Lagerstroemia speciosa, Morinda citrifolia, towards various proteins, eg, tyrosine kinase,35 estrogen receptor-alpha, progesterone receptor, and epidermal growth factor receptor,36 alpha-glucosidase and alpha-amylase,24 and pyruvate carboxylase,37 have been reported with tolerable binding energy.
SIRT1, a nicotinamide adenine dinucleotide-dependent deacetylase, is associated with various metabolic pathways regulating the energetic state of the cell, including enhanced mitochondrial biogenesis, insulin sensitivity, and a lower incidence of age-related diseases.21 The activation of SIRT1 was thought to interfere with the lipogenesis process.6–8 The present molecular docking study revealed that both beta-sitosterol and stigmasterol interact with SIRT1 by building pi-alkyl hydrophobic interactions with Ile223, Ile227, and Pro212 residues in the allosteric region of the alpha-helices of the enzyme. These interactions are similar to those of 4TQ and resveratrol. The binding energies of these sterol compounds confirm that they are all active and effectively occupy the allosteric sites. The pi–alkyl hydrophobic interactions may contribute to stabilizing the conformation of the phytosterols. However, the known allosteric activators of SIRT1, 4TQ, and resveratrol demonstrated a stronger affinity (lower binding energy) due to their hydrogen bond with Glu230, which is not shown by the phytosterols.
The crystal structure of human SIRT1 in complex with a small molecule sirtuin-activating compound was previously announced. It was reported that the small molecule sirtuin-activating compound directly interacts with the N-terminal domain of the enzyme by occupying a narrow pocket far from the catalytic domain. Ile223, Ile227, Pro212, and Glu230 are important amino acid residues that assemble a binding pocket in the N-terminal domain of alpha-helices which plays a pivotal role in the allosteric activation mechanism of SIRT1.21 Moreover, in the activated SIRT1 conformation, another activator namely SCIC2 interacts with the hydrophobic residues Leu202, Leu206, and Ile223, while the benzyl moiety is directed to the Glu230 side chain.38
A previous study proposed that the binding of resveratrol to SIRT1 activates a conformational change in the enzyme which permits tighter fluorophore binding in the context of the entire peptide substrate.39 Glu230 residue is another important amino acid in SIRT1 which is responsible for the conformational stability of SIRT1 when bound to the substrate.40 Our study was also in agreement with a previous study that described the interaction of stigmasterol with Pro212 may correlate with the increase of SIRT1 expression in SH-SY5Y cells.41 The binding of a compound, eg, resveratrol, to important residues in the N-terminal domain has the potential to increase SIRT1 activity by closing the N-terminal domain.42
AMPK is an enzyme that, when present in its active form, inhibits the process of lipid synthesis. Thus, compounds that bind to AMPK might activate the protein through several mechanisms. The first is by increasing the activity of the serine-threonine liver kinase B1 (LKB1), a protein that phosphorylates AMPK; the second is by increasing the phosphorylation rate in the catalytic domain of the alpha subunit (Thr172 residue), and the last is by inhibiting the action of a protein that causes AMPK dephosphorylation.43–46 The crystal structure of the full-length human AMPK complex co-crystallized with A-769662 revealed that A-769662 resides in a site between the alpha-subunit and the carbohydrate-binding module of the beta-subunit, located separately from the adenine nucleotide-binding sites on the AMPK gamma-subunit.22
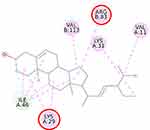
This study verified that the binding mode of β-sitosterol and stigmasterol with AMPK is similar to that of A-769662, that is by building hydrogen bonds with Lys29 and Arg83 residues. It was recorded that A-769662 binds to Lys29, Arg83, Asp88, and Ser108 residues. Meanwhile, metformin only interacts with Arg83. Our results are in agreement with an earlier study, that AMPK activators interact with Lys29, Arg83, Asp88, and Ser108.47 Similarly, a previous study proposed that AMPK activators should possess a favorable feature, which is the formation of a salt bridge with Lys29.48 Furthermore, a former in vitro study has shown the potential of beta-sitosterol as an AMPK activator, by increasing the expression of AMPK in L6 myotube cells.49
Thienopyridone (synonym A-769662) and metformin were used as standards because these two drugs are well-known as AMPK activators.43,44 Metformin stimulates AMPK in the liver cells and eventually reduces the activity of acetyl-CoA carboxylase, induces fatty acid oxidation, and suppresses the expression of lipogenic enzymes. Activation of AMPK by metformin triggers the suppression of SREBP-1 expression. It was confirmed that in the isolated rat skeletal muscles, this biguanide oral antihyperglycemic drug induces glucose uptake related to AMPK activation.45 A-769662 is a drug manufactured by Abbott Laboratories that generates allosteric activation of purified AMPK in cell-free assays and blocks fatty acid synthesis in rat hepatocytes.43 It was suggested that A-769662 and AMP occupy distinct binding sites on the AMPK complex, thus exhibiting disparate mechanisms of activation.46
The pharmacophores of both beta-sitosterol and stigmasterol comprise one hydrogen bond donor (the hydroxyl group attached to C-3 in ring A) and three hydrophobic groups (the aromatic rings A, B, and C) which is in accordance with a previous molecular docking study of phytosterols towards α-amylase, α-glucosidase, and DPP-IV that reported similar pharmacophoric features.23 A pharmacophore is an entity of chemical features that identify a particular mode of action of a ligand in the active site of a macromolecule. The most important pharmacophoric features are hydrogen bond acceptors, hydrogen bond donors, hydrophobicity, positively and negatively ionizable groups, aromatic groups, and metal coordinating areas.50,51 Hydrogen bonds are essential in regulating the specificity of the binding of a drug to its macromolecule target. These bonds are weak, easily be broken by the increase in temperature, and distance change, and represent a special type of dipole–dipole attraction between two molecules.52 β-sitosterol and stigmasterol might activate SIRT1 and AMPK thus preventing the generation of fatty liver and improving lipid disorders such as NAFLD. Previous studies showed that the activation of SIRT1 and AMPK inhibited the formation of lipogenesis enzymes resulting in the inhibition of lipogenesis.43
Our current work does not involve in vitro or in vivo studies in animal models, which reflects its limitations. Therefore, further explorations are needed to elucidate a complete picture of the effect of beta-sitosterol and stigmasterol on SIRT1 and AMPK in NAFLD conditions.
Conclusion
The present work explored the ethanol extract of a plant collected from a rainforest in East Kalimantan, Indonesia, Lygodium microphyllum, which is one of the fern-type plants with an abundance of pharmacological activities. The benefit of L. microphyllum was rooted in many active metabolites contained therein. Our study revealed that the ethanol extract of L. microphyllum (ELM) contained steroids, phenols, flavonoids, alkaloids, and saponins. ELM contains high total phenolic compounds (126 mg GAE/g), total sterols (954.04 µg/g), and beta-sitosterol contained in the extract is 283.55 µg/g. Our present study is the first that reported the amount of beta-sitosterol in ELM as well as the moderate toxicity of ELM (LC50 of 203.704 ppm) towards Artemia salina. Thus, a molecular docking simulation was employed to study whether beta-sitosterol and stigmasterol, bind to the allosteric site residues of sirtuin 1 (SIRT1) and adenosine monophosphate-activated protein kinase (AMPK), in the same manner as those of the protein activators. SIRT1 and AMPK are two important proteins in the lipogenesis mechanism. We declare that this is the first report of the binding mode of phytosterols of L. microphyllum towards SIRT1 and AMPK. Both β-sitosterol and stigmasterol could occupy the allosteric-binding site of SIRT1 and AMPK by binding to Ile223, Ile227, Pro212, and Glu230 of SIRT1 and Lys29 and Arg83 of AMPK, similar to the proteins’ known activators. These findings provide evidence that ELM can be further explored for its pharmacology activities for the discovery of plant-based anti-dyslipidemia drug candidates. Nonetheless, additional studies are still required to elucidate how SIRT1 and AMPK are activated by β-sitosterol and stigmasterol.
Acknowledgments
The authors thank (1) the Faculty of Pharmacy, Mulawarman University, (2) the Directorate of Higher Education of the Ministry of Education and Culture for funding the research, and (3) the Rector of Padjadjaran University via the Directorate of Research and Community Engagement.
Funding
This research is funded by the Directorate of Higher Education of the Ministry of Education and Culture via the Doctoral Dissertation Grant no. 044/E5/PG.02.00.PL/2023 and 031/E5/PG.02.00.PL/2023. The APC is funded by Padjadjaran University via the Directorate of Research and Community Engagement.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Iqbal U, Perumpail BJ, Akhtar D, et al. The epidemiology, risk profiling, and diagnostic challenges of non-alcoholic fatty liver disease. Medicines. 2019;6(1):41. doi:10.3390/medicines6010041
2. Anggreini P, Kuncoro H, Sumiwi SA, et al. Role of the AMPK/SIRT1 pathway in non-alcoholic fatty liver disease. Mol Med Rep. 2023;27(2):35. doi:10.3892/mmr.2022.12922
3. Yoo JJ, Kim W, Kim MY, et al. Recent research trends and updates on non-alcoholic fatty liver disease. Clin Mol Hepatol. 2019;25(1):1. doi:10.3350/cmh.2018.0037
4. Singh CK, Chhabra G, Ndiaye MA, et al. The role of sirtuins in antioxidant and redox signaling. Antioxid Redox Signal. 2018;28(8):643. doi:10.1089/ars.2017.7290
5. Rahman S, Islam R. Mammalian Sirt1: insights on its biological functions. Cell Commun Signal. 2011;9(1):11. doi:10.1186/1478-811X-9-11
6. Li S, Qian Q, Ying N, et al. Activation of the AMPK-SIRT1 pathway contributes to the protective effects of salvianolic acid a against lipotoxicity in hepatocytes and NAFLD in mice. Front Pharmacol. 2020;11:560905. doi:10.3389/fphar.2020.560905
7. Ding RB, Bao J, Deng CX. Emerging roles of SIRT1 in fatty liver diseases. Int J Biol Sci. 2017;13(7):852–867. doi:10.7150/ijbs.19370
8. Schug TT, Li X. Sirtuin 1 in lipid metabolism and obesity. Ann Med. 2011;43(3):198–211. doi:10.3109/07853890.2010.547211
9. Wang Q, Liu S, Zhai A, et al. AMPK-mediated regulation of lipid metabolism by phosphorylation. Biol Pharm Bull. 2018;41(7):985–993. doi:10.1248/bpb.b17-00724
10. Hardie DG, Ross FA, Hawley SA. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol. 2012;13(4):251–262. doi:10.1038/nrm3311
11. Ha JH, Jang J, Chung SI, et al. AMPK and SREBP-1c mediate the anti-adipogenic effect of β-hydroxyisovalerylshikonin. Int J Mol Med. 2016;37(3):816–824. doi:10.3892/ijmm.2016.2484
12. Fouqueray P, Bolze S, Dubourg J, et al. Pharmacodynamic effects of direct AMP kinase activation in humans with insulin resistance and non-alcoholic fatty liver disease: a phase 1b study. Cell Rep Med. 2021;2(12):100474. doi:10.1016/j.xcrm.2021.100474
13. Frasinariu O, Serban R, Trandafir LM, et al. The role of phytosterols in nonalcoholic fatty liver disease. Nutrients. 2022;14(11):2187. doi:10.3390/nu14112187
14. Alam MM, Emon NU, Alam S, et al. Assessment of pharmacological activities of Lygodium microphyllum Cav. leaves in the management of pain, inflammation, pyrexia, diarrhea, and helminths: in vivo, in vitro and in silico approaches. Biomed Pharmacother. 2021;139(5):111644. doi:10.1016/j.biopha.2021.111644
15. Kuncoro H, Rijai L.
16. Dumolt JH, Rideout TC. The lipid-lowering effects and associated mechanisms of dietary phytosterol supplementation. Curr Pharm Des. 2017;23(34):5077–5085. doi:10.2174/1381612823666170725142337
17. Kuncoro H, Farabi K, Rijai L. Steroids and isoquercetin from. Lygodium microphyllum. J Appl Pharm Sci. 2017;7(11):136–141.
18. Siddiqui N, Rauf A, Latif A, et al. Spectrophotometric determination of the total phenolic content, spectral and fluorescence study of the herbal Unani drug Gul-e-Zoofa (Nepeta bracteata Benth). J Taibah Univ Med Sci. 2017;12(4):360–363. doi:10.1016/j.jtumed.2016.11.006
19. Matysik E, Woźniak A, Paduch R, et al. The new TLC method for separation and determination of multicomponent mixtures of plant extracts. J Anal Methods Chem. 2016;2016:1–6. doi:10.1155/2016/1813581
20. Lestari MS, Himawan T, Abadi AL, et al. Toxicity and phytochemistry test of methanol extract of several plants from Papua using brine shrimp lethality test (BSLT). J Chem Pharm Res. 2015;7(4):866–872.
21. Dai H, Case AW, Riera TV, et al. Crystallographic structure of a small molecule SIRT1 activator-enzyme complex. Nat Commun. 2015;6(1):7645. doi:10.1038/ncomms8645
22. Xiao B, Sanders MJ, Carmena D, et al. Structural basis of AMPK regulation by small molecule activators. Nat Commun. 2013;4(1):3017. doi:10.1038/ncomms4017
23. Lolok N, Ramadhan DFS, Sumiwi SA, et al. Molecular docking of β-sitosterol and stigmasterol isolated from Morinda citrifolia with α-amylase, α-glucosidase, dipeptidyl peptidase-IV, and peroxisome proliferator-activated receptor-γ. Rasayan J Chem. 2022;15(1):20. doi:10.31788/RJC.2022.1516646
24. Lolok N, Ramadhan DFS, Sumiwi SA, et al. Molecular dynamics study of stigmasterol and beta-sitosterol of Morinda citrifolia L. towards α-amylase and α-glucosidase. J Biomol Struct Dyn. 2023:1–4. doi:10.1080/07391102.2023.2243519
25. Gylling H, Plat J, Turley S, et al. Plant sterols and plant stanols in the management of dyslipidemia and prevention of cardiovascular disease. Atherosclerosis. 2014;232(2):346–360. doi:10.1016/j.atherosclerosis.2013.11.043
26. AbuMweis SS, Marinangeli CP, Frohlich J, et al. Implementing phytosterols into medical practice as a cholesterol-lowering strategy: an overview of efficacy, effectiveness, and safety. Can J Cardiol. 2014;30(10):1225–1232. doi:10.1016/j.cjca.2014.04.022
27. Gylling H, Simonen P. Phytosterols, phytostanols, and lipoprotein metabolism. Nutrients. 2015;7(9):7965–7977. doi:10.3390/nu7095374
28. Ras RT, Fuchs D, Koppenol WP, et al. The effect of a low-fat spread with added plant sterols on vascular function markers: results of the Investigating Vascular Function Effects of Plant Sterols (INVEST) study. Am J Clin Nutr. 2015;101(4):733–741. doi:10.3945/ajcn.114.102053
29. Reymon R, Sofyan S, Yodha AW, Musdalipah M. The toxicity of Meistera chinensis rhizome fraction by shrimp larvae with BSLT method. Nat Sci. 2021;10(2):53–58.
30. Ayaz M, Sadiq A, Wadood A, et al. Cytotoxicity and molecular docking studies on phytosterols isolated from Polygonum hydropiper L. Steroids. 2019;141:30–35. doi:10.1016/j.steroids.2018.11.005
31. Raju L, Lipin R, Eswaran R. Identification, ADMET evaluation and molecular docking analysis of phytosterols from Banaba (Lagerstroemia speciosa (L.) Pers) seed extract against breast cancer. In Silico Pharmacol. 2021;9(1):43. doi:10.1007/s40203-021-00104-y
32. Uttu AJ, Sallau MS, Ibrahim H, Iyun ORA. Isolation, characterization, and docking studies of campesterol and β-sitosterol from Strychnos innocua (Delile) root bark. J Taibah Univ Med Sci. 2022;18(3):566–578. doi:10.1016/j.jtumed.2022.12.003
33. Sujana D, Sumiwi SA, Saptarini NM, et al. ADMET prediction and molecular docking simulation of phytoconstituents in Boesenbergia rotunda rhizome with the effector caspases to understand their protective effects. Rasayan J Chem. 2022;15(4):2401. doi:10.31788/RJC.2022.1547011
34. Bordone L, Cohen D, Robinson A, et al. SIRT1 transgenic mice show phenotypes resembling calorie restriction. Aging Cell. 2007;6(6):759–767. doi:10.1111/j.1474-9726.2007.00335.x
35. Ding S, Jiang J, Zhang G, et al. Resveratrol and caloric restriction prevent hepatic steatosis by regulating SIRT1-autophagy pathway and alleviating endoplasmic reticulum stress in high-fat diet-fed rats. PLoS One. 2017;12(8):8. doi:10.1371/journal.pone.0183541
36. You M, Jogasuria A, Taylor C, et al. Sirtuin 1 signaling and alcoholic fatty liver disease. Hepatobiliary Surg Nutr. 2015;4(2):88–100. doi:10.3978/j.issn.2304-3881.2014.12.06
37. Ramirez T, Li YM, Yin S, et al. Aging aggravates alcoholic liver injury and fibrosis in mice by downregulating sirtuin 1 expression. J Hepatol. 2017;66(3):601–609. doi:10.1016/j.jhep.2016.11.004
38. Scisciola L, Sarno F, Carafa V, et al. Two novel SIRT1 activators, SCIC2 and SCIC2.1, enhance SIRT1-mediated effects in stress response and senescence. Epigenetics. 2020;15(6–7):664–683. doi:10.1080/15592294.2019.1704349
39. Borra MT, Smith BC, Denu JM. Mechanism of human SIRT1 activation by resveratrol. J Biol Chem. 2005;280(17):17187. doi:10.1074/jbc.M501250200
40. Feddal S, Bouakouk Z, Meyar M, et al. Atomic 3D-QSAR study based on pharmacophore modeling of resveratrol derivatives as selective COX-2 inhibitors. Med Chem Res. 2017;26(6):1259. doi:10.1007/s00044-017-1830-0
41. Pratiwi R, Nantasenamat C, Ruankham W, et al. Mechanisms and neuroprotective activities of stigmasterol against oxidative stress-induced neuronal cell death via Sirtuin family. Front Nutr. 2021;8:648995. doi:10.3389/fnut.2021.648995
42. Hou X, Rooklin D, Fang H, et al. Resveratrol serves as a protein-substrate interaction stabilizer in human SIRT1 activation. Sci Rep. 2016;6(1):38186. doi:10.1038/srep38186
43. Cool B, Zinker B, Chiou W, et al. Identification and characterization of a small molecule AMPK activator that treats key components of type 2 diabetes and the metabolic syndrome. Cell Metab. 2006;3(6):403–416. doi:10.1016/j.cmet.2006.05.005
44. Banerjee J, Bruckbauer A, Thorpe T, et al. Biphasic effect of sildenafil on energy sensing is mediated by phosphodiesterases 2 and 3 in adipocytes and hepatocytes. Int J Mol Sci. 2019;20(12):2992. doi:10.3390/ijms20122992
45. Zhou G, Myers R, Li Y, et al. Role of AMP-activated protein kinase in mechanism of metformin action. J Clin Invest. 2001;108(8):1167–1174. doi:10.1172/JCI13505
46. Sanders MJ, Ali ZS, Hegarty BD, et al. Defining the mechanism of activation of AMP-activated protein kinase by the small molecule A-769662, a member of the thienopyridone family. J Biol Chem. 2007;282(45):32539–32548. doi:10.1074/jbc.M706543200
47. Li Y, Peng J, Li P, et al. Identification of potential AMPK activator by pharmacophore modeling, molecular docking and QSAR study. Comput Biol Chem. 2019;79:165–176. doi:10.1016/j.compbiolchem.2019.02.007
48. Hao J, Yang Z, Li J, et al. Discovery of natural adenosine monophosphate-activated protein kinase activators through virtual screening and activity verification studies. Mol Med Rep. 2021;23(3):203. doi:10.3892/mmr.2021.11842
49. Hwang SL, Kim HN, Jung HH, et al. Beneficial effects of beta-sitosterol on glucose and lipid metabolism in L6 myotube cells are mediated by AMP-activated protein kinase. Biochem Biophys Res Commun. 2008;377(4):1253–1258. doi:10.1016/j.bbrc.2008.10.136
50. Daisy P, Singh SK, Vijayalakshmi P, et al. A database for the predicted pharmacophoric features of medicinal compounds. Bioinformation. 2011;6(4):167–168. doi:10.6026/97320630006167
51. Giordano D, Biancaniello C, Argenio MA, et al. Drug design by pharmacophore and virtual screening approach. Pharmaceuticals. 2022;15(5):646. doi:10.3390/ph15050646
52. Wade RC, Goodford PJ. The role of hydrogen bonds in drug binding. Prog Clin Biol Res. 1989;289:433–444.
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.