Back to Journals » Infection and Drug Resistance » Volume 17
Isolation and Characterization of Cholera Toxin Gene-Positive Vibrio cholerae Non-O1/Non-O139 Isolated from Urinary Tract Infection: A Case Report
Authors Aljindan R, Allahham R, Alghamdi R, Alhabib I , AlNassri S , Alkhalifa W, Diab A, Alomar A, Yamani L , Elhadi N
Received 3 January 2024
Accepted for publication 16 March 2024
Published 21 March 2024 Volume 2024:17 Pages 1147—1152
DOI https://doi.org/10.2147/IDR.S456654
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Reem Aljindan,1 Reema Allahham,2 Rana Alghamdi,2 Ibrahim Alhabib,2 Samia AlNassri,3 Wala Alkhalifa,4 Asim Diab,1 Amer Alomar,2 Lamya Yamani,2 Nasreldin Elhadi2
1Department of Microbiology, College of Medicine, Imam Abdulrahman Bin Faisal University, Dammam, Kingdom of Saudi Arabia; 2Department of Clinical Laboratory Science, College of Applied Medical Sciences, Imam Abdulrahman Bin Faisal University, Dammam, Kingdom of Saudi Arabia; 3Department of Infection Control, King Fahad Hospital of the University, Alkhobar, Kingdom of Saudi Arabia; 4Department of Microbiology, Comprehensive Screening Center, Eastern Health Cluster, Dammam, Kingdom of Saudi Arabia
Correspondence: Nasreldin Elhadi, Department of Clinical Laboratory Science, College of Applied Medical Sciences, Imam Abdulrahman Bin Faisal University, P.O. Box 2435, Dammam, 31441, Kingdom of Saudi Arabia, Email [email protected]
Background: Urinary tract infection (UTI) caused by V. cholerae is rare and less common. V. cholerae is a Gram-negative bacterium motile using single polar flagellum and, originally, is a waterborne microbe found in aquatic and estuarine environments. Toxigenic V. cholerae is well-known as a causative agent of acute and excessive watery diarrhea after ingesting food and water contaminated with this bacterium.
Case Presentation: A 27-year-old male patient presented to the emergency department on 17th July 2021 with burning micturition, normal vital signs, and no fever, vomiting, or diarrhea. In 2017, the patient complained of short stature and vitamin D deficiency. He was on human growth hormone from January 2018 till October 2019. The diagnosis was V. cholerae Non-O1/non-O139 urinary tract infection (UTI). Considering a urinary tract infection, empirical treatment with Lornoxicam and Ciprofloxacin was initiated, while the result of urine culture was still pending. The patient was discharged on the same day and without any complications.
Conclusion: V. cholerae non-O1/non-O139 is primarily a marine inhabitant and is associated with sporadic cases resulting in cholera-like diarrhea after consumption of contaminated seafood and exposure to seawater. Extraintestinal infection associated with this bacterium should no longer be ignored as this change in the behavior of cholera bacteria mechanism of pathogenicity might be related to some associated virulence genes.
Keywords: non-O1/non-O139 V. cholerae, virulence genes, extraintestinal infection, ERIC-PCR
Background
Vibrio cholerae is Gram negative-bacteria that habituate marine and estuarine environments. Cholera infection remains a potential public health problem, and it is not restricted to underdeveloped countries but has been seen in developing countries.1,2 Toxigenic strains of V. cholerae are responsible for causing acute dehydrating diarrhea that occurs in an epidemic form in many developing countries. V. cholerae is currently classified into more than 200 serogroups, and the toxigenic strains producing cholera toxin, that are responsible for epidemics and pandemics, belong to serogroups O1 and O139.3 Non-O1/non-O139 V. cholerae (NOVC), which does not agglutinate with O1 and O139 antisera, could also cause sporadic cases of gastroenteritis and other extraintestinal infections such as bacteremia, keratitis and wound infections.2,4–7
The cholera toxin (CT) is the major secreted toxin by V. cholerae serogroup O1 and O139 and is responsible for severe watery diarrhea, which could lead to dehydration and death if left untreated.8 The other virulence factor is toxin-coregulated pilus (TCP), which is required for small intestine colonization.8 It has been documented that NOVC isolates are lacking CT and TCP, therefore, they establish infections through other virulence factors such as zonula occludens toxin (zot), accessory cholera enterotoxin (ace), and hemolysin (hlyA).9,10 Mechanism of extraintestinal infections caused by NOVC is not yet fully understood, but it has been accepted that more than 85% of NOVCs have a capsule that may be involved in combination with other virulence factors in causing extraintestinal infections.11 We here present a case of non-O1/non-O139 V. cholerae (NOVC) causing a urinary tract infection in a 27-year-old male patient. To our knowledge, this is the first case report in Saudi Arabia reporting the occurrence of urinary tract infection caused by V. cholerae non-O1/non-O139.
Case Presentation
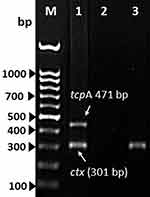
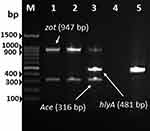
A 27-year-old Saudi male from Al-Khobar City in the Eastern Province of Saudi Arabia presented to the emergency department at King Fahd Hospital of the University (KFHU) on 17th July 2021 with burning micturition. The patient had a history of swimming the day before in Halfmoon beach area situated on the coast of the Arabian Gulf. His urine analysis revealed negative nitrite, 3+ leukocytes, numerous white blood cells (30–50), red blood cells more than 200, and 2+ bacteria. Urine culture was done on blood agar plate and revealed ß-hemolytic colonies. Bacterial colonies were identified as V. cholerae using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS), Biomèrieux, France. The identification by MALDI-TOF MS was verified on thiosulfate citrate bile salts sucrose (TCBS) agar. Colonies on TCBS revealed sucrose fermenting yellow colonies of typical characteristic of V. cholerae, which was further confirmed using Vitek2 (Biomèrieux, France). The serological of isolate was determined by slide agglutination test using V. cholerae O1 and V. cholerae O139 polyvalent antisera (MAST ASSURE Antiserum Vibrio cholerae polyvalent, UK). The isolate gave a negative result with both polyvalent antiserum and reported as non-O1/non-O139 V. cholerae strain according to the manufacturer’s instruction. Antimicrobial susceptibility testing was carried out using Vitek2 (Biomèrieux, France), and the isolate was resistant to ampicillin and susceptible to ciprofloxacin and trimethoprim-sulfamethoxazole. The isolate was further confirmed as V. cholerae by PCR targeting the V. cholerae species specific toxR (Figure 1A) and ompU genes (Figure 1B) and strain tested positive for both genes. Furthermore, the strain was tested positive for major cholera toxin (ctx) gene and lack the TCP gene which is required for colonization (Figure 2). Multiplex-PCR analysis revealed V. cholerae strain was harboring gene encoding hemolysin (hlyA) which play significant role in disease mechanism and invasion, while it was lacking the zonular occludens toxin (zot) and accessory colonization enterotoxin (ace) genes (Figure 3). ERIC-PCR was used as genomic fingerprinting for determination of clonality and relatedness of isolated V. cholerae strain from urine with three V. cholerae control strains (Figure 4). Based on analyzed result of ERIC fingerprint patterns using UPGMA and Pearson correlation coefficient, isolated V. cholerae strains share 90% genetic similarity with the control strain of V. cholerae O139 ATCC 5139 (Figure 4).
Discussion
V. cholerae is a Gram-negative bacterium and highly motile by the means of single polar flagellum, whose natural habitat is usually costal water and marine environment. Among more than 200 serogroups that have been identified in V. cholerae, only O1 and O139 are related to cholera pandemic outbreak. V. cholerae, particularly serogroups O1 and O139, is well-known for causing severe watery diarrhea, a potentially life-threatening illness if left untreated. Transmission typically occurs through contaminated water and food. Additionally, non-O1/non-O139 V. cholerae and other non-cholera Vibrio species, such as V. parahaemolyticus, V. vulnificus, V. alginolyticus, and V. fluvialis, can cause infections, usually acquired through consumption of contaminated seafood or exposure to seawater.12,13
Both O1 and O139 serogroup produce cholera toxin (ctx) and toxin co-regulated pilus (TCP), which are the major virulence factors associated with V. cholerae infection. However, V. cholerae non-O1/non-O139 strains are associated with sporadic cases of diarrhea and extra-intestinal infections, and some of these strains have been reported to possess virulence factors encoding zot, ace and hlyA.9,10,14 In recent years, cases of extra-intestinal infections caused by V. cholerae non-O1/non-O139 are on the rise and have been reported from different parts of the world causing infections such as septicemia, meningitis, cellulitis, and keratitis.4,5,15,16 It is well-known that V. cholerae is the causative agent of cholera, a gastrointestinal disease, after ingestion of contaminated food or water, where the infection leads to voluminous watery diarrhea causing extreme dehydration. Non-O1/non-O139 may cause a cholera-like illness similar to pandemic V. cholerae serogroup O1 and O139, however, distinct pathways of pathogenesis mechanisms may exist.14 Some non-O1/non-O139 strains may harbor cholera toxin gene or produce similar choleragen that have been seen in most diarrheal cases. Both serogroups of pandemic V. cholerae O1 and O139 are a non-invasive intestinal bacterial pathogens and have never been isolated from body fluids other than fecal stool specimen, because these serogroups do not breach the mucosal surface of the intestine. In contrast, V. cholerae non-O1/non-O139 strains are potentially invasive and implicated in invasive infections leading to septicemia and other extra-intestinal infections.4,17
In this case study, reporting the isolated strain harboring the ctxA gene encoding the A subunit of CT is considered important. Usually, strains of V. cholerae non-O1/non-O139 are lacking the pathogenicity island of CT and TCP but harbor other virulence factors such as heat stable enterotoxin, hemolysin, and zonula occludens toxin. The association of these virulence gene factors with extra-intestinal infections has been documented by several studies.18,19
In Saudi Arabia, cases that are unrelated to international travel of extra-intestinal V. cholerae non-O1/non-O139 infections has been reported by several studies.6,20–23 V. cholerae strain in the current case was investigated and compared with control strains of V. cholerae O1 Ogawa (Classical biotype), V. cholerae O1 Inaba (Classical biotype) and V. cholerae O139 (Bengal) using ERIC-PCR to find the possibility of epidemiological link. Therefore, urine strain of V. cholerae was grouped with control strain of V. cholerae O139 and shared 90% genetic similarity. Further molecular epidemiology investigation of this strain and other strains of V. cholerae non-O1/non-O139 isolated from other intestinal and extra-intestinal infections including coastal water and sewerage on future studies is essential and should be conducted to identify the potential sources and route of infections. In addition, the possible effects of global warming on infectious diseases may broaden the endemic area of cholera infection since V. cholerae growth is abundance in warm waters of tropical and subtropical climate.
Conclusion
This study suggests that more attention should be directed towards the potential for future endemicity of V. cholerae in areas where few cases are currently reported. The current report emphasizes that V. cholerae non-O1/non-O139 should be suspected in patients who present symptoms of urinary tract infection. Moreover, similar infections may increase in the future in an area where V. cholerae is not endemic such as Saudi Arabia due to the climate change effects on infectious diseases. From reviewed case reports in the literature, several studies reported rare and unusual cases of extra-intestinal infections associated with V. cholerae non-O1/non-O139; therefore, more intensive studies are needed to understand this change in the behavior of V. cholerae non-O1/non-O139 from sporadic intestinal infections to broad extra-intestinal infections. However, this higher likelihood of a potential increase in V. cholerae non-O1/non-O139 extra-intestinal infection cases is reported globally, and there is a possibility that this increase could be linked to the increased number of vulnerable patients who are immunocompromised and to the climate change. Clinical awareness in hospitals and laboratories is required to ensure optimal patient outcomes by considering V. cholerae non-O1/non-O139 might cause urinary tract infections after swimming exposure to seawater. To our knowledge, this is the first report of V. cholerae non-O1/non-O139 (ctx+) isolated from urine in Saudi Arabia.
Data Sharing Statement
The original contributions presented in the study are included in the article. Further inquiries can be directed to the corresponding author.
Ethics Approval
The patient had given its written informed consent for the use of his personal and medical information for the publication of this study. This study was approved by the Institutional Review Board (IRB) of Imam Abdulrahman Bin Faisal University (IRB approval number: IRB-2022-03-220).
Funding
The authors received no extramural funding for the study.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Faruque SM, Nair GB. Molecular ecology of toxigenic Vibrio cholerae. Microbiol Immunol. 2002;46(2):59–66. doi:10.1111/j.1348-0421.2002.tb02659.x
2. Montero DA, Vidal RM, Velasco J, et al. Vibrio cholerae, classification, pathogenesis, immune response, and trends in vaccine development. Front Med. 2023;10(May). doi:10.3389/fmed.2023.1155751
3. Sampaio A, Silva V, Poeta P, Aonofriesei F. Vibrio spp: life strategies, ecology, and risks in a changing environment. Diversity. 2022;14(2):1–26. doi:10.3390/d14020097
4. Chowdhury G, Joshi S, Bhattacharya S, et al. Extraintestinal infections caused by non-toxigenic Vibrio cholerae non-O1/non-O139. Front Microbiol. 2016;7(FEB):1–5. doi:10.3389/fmicb.2016.00144
5. Chen WD, Lai LJ, Hsu WH, Huang TY. Vibrio cholerae non-O1 - The first reported case of keratitis in a healthy patient. BMC Infect Dis. 2019;19(1):1–5. doi:10.1186/s12879-019-4475-4
6. Abdelhafiz TA, Alnimr AM, Alabduljabbar AM, et al. Non O1 Vibrio cholerae as a cause of bacteremic lower limb cellulitis: a case report. Int J Surg Case Rep. 2019;64:62–65. doi:10.1016/j.ijscr.2019.09.020
7. Deshayes S, Daurel C, Cattoir V, Parienti JJ, Quilici ML, de La Blanchardière A. Non-O1, non-O139 Vibrio cholerae bacteraemia: case report and literature review. Springerplus. 2015;4(1):1–9. doi:10.1186/s40064-015-1346-3
8. Faruque SM, Albert MJ, Mekalanos JJ. Epidemiology, Genetics, and Ecology of Toxigenic Vibrio cholerae. Microbiol Mol Biol Rev. 1998;62(4):1301–1314. doi:10.1128/mmbr.62.4.1301-1314.1998
9. Singh DV, Matte MH, Matte GR, et al. non-O139 strains: clonal relationships between clinical and environmental isolates. Society. 2001;67(2):910–921. doi:10.1128/AEM.67.2.910
10. Pérez-Reytor D, Jaña V, Pavez L, Navarrete P, García K. Accessory toxins of vibrio pathogens and their role in epithelial disruption during infection. Front Microbiol. 2018;9(SEP):1–11. doi:10.3389/fmicb.2018.02248
11. Chen Y, Bystricky P, Adeyeye J, et al. The capsule polysaccharide structure and biogenesis for non-O1 Vibrio cholerae NRT36S: genes are embedded in the LPS region. BMC Microbiol. 2007;7:1–15. doi:10.1186/1471-2180-7-20
12. Baker-Austin C, Oliver JD, Alam M, et al. Vibrio spp. infections. Nat Rev Dis Primers. 2018;4(1):1–19. doi:10.1038/s41572-018-0005-8
13. Hecht J, Borowiak M, Fortmeier B, et al. Case Report: vibrio fluvialis isolated from a wound infection after a piercing trauma in the Baltic Sea. Access Microbiol. 2022;4(1). doi:10.1099/acmi.0.000312
14. Igere BE, Okoh AI, Nwodo UU. Non-serogroup O1/O139 agglutinable Vibrio cholerae: a phylogenetically and genealogically neglected yet emerging potential pathogen of clinical relevance. Arch Microbiol. 2022;204(6):1–28. doi:10.1007/s00203-022-02866-1
15. Hao Y, Wang Y, Bi Z, et al. A case of non-O1/non-O139 Vibrio cholerae septicemia and meningitis in a neonate. Int J Infect Dis. 2015;35:117–119. doi:10.1016/j.ijid.2015.05.004
16. Clark R, Bracy W, Hanna B, Love GL. Case report: vibrio cholerae non-O1 infection presenting as localized cellulitis. Am J Med Sci. 1989;298(5):328–330. doi:10.1097/00000441-198911000-00010
17. Chen Y, Tang H, Chao C, Lai C. Clinical Manifestations of Non-O1 Vibrio cholerae Infections. PLoS One. 2015;4–11. doi:10.1371/journal.pone.0116904
18. Leong LEX, Gordon DL, Rogers B. Draft genome sequence of a non-O1/O139 Vibrio cholerae strain isolated from a patient presenting with dysuria. Microbiol Resour Announc. 2018;2018:1–2.
19. Zhao Y, He T, Tu B, et al. Death in a farmer with underlying diseases carrying Vibrio cholerae non-O1/non-O139 producing zonula occludens toxin. Int J Infect Dis. 2022;120:83–87. doi:10.1016/j.ijid.2022.04.020
20. Eltahawy AT, Jiman-Fatani AA, Al-Alawi MM. A fatal non-01 Vibrio cholerae septicemia in a patient with liver cirrhosis. Saudi Med J. 2004;25(11):1730–1731.
21. Issa H. A case of O1 vibrio cholera bacteremia and primary peritonitis in a patient with liver cirrhosis. Gastroenterol Res. 2009;2(6):358–360. doi:10.4021/gr2009.12.1331
22. Kaki R, El-Hossary D, Jiman-Fatani A, Al-Ghamdi R. Non-O1/non-O139 Vibrio cholerae septicaemia in a Saudi man: a case report. JMM Case Reports. 2017;4(2):1–6. doi:10.1099/jmmcr.0.005077
23. Nagamani R, Al Momen HAM. Non O1 Vibrio cholera: an emerging pathogen in blood?-A review and report of cases from a regional laboratory at the Eastern Province in Saudi Arabia. MRIMS J Heal Sci. 2017;5(3):109. doi:10.4103/2321-7006.302676
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.