Back to Journals » Journal of Inflammation Research » Volume 17
Investigation of the miRNA-mRNA Regulatory Circuits and Immune Signatures Associated with Bronchopulmonary Dysplasia
Authors Li S, Liang S, Xie S, Chen H, Huang H, He Q, Zhang H, Wang X
Received 6 November 2023
Accepted for publication 29 February 2024
Published 5 March 2024 Volume 2024:17 Pages 1467—1480
DOI https://doi.org/10.2147/JIR.S448394
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Ning Quan
Sen Li,1 Shuling Liang,2 Shunyu Xie,1 Haixia Chen,1 Haoying Huang,1 Qixin He,3 Huayan Zhang,4,5 Xiaohui Wang1
1Guangzhou Women and Children’s Medical Center, State Key Laboratory of Respiratory Disease and Guangzhou Medical University, Guangzhou, Guangdong Province, People’s Republic of China; 2Guangdong Provincial Research Center for Child Health, Guangzhou Women and Children’s Medical Center, Guangzhou Medical University, Guangzhou, Guangdong Province, People’s Republic of China; 3Sun Yat-sen University Cancer Center, Guangzhou, Guangdong Province, People’s Republic of China; 4Division of Neonatology and Center for Newborn Care, Guangzhou Women and Children’s Medical Center, Guangzhou, Guangdong Province, People’s Republic of China; 5Division of Neonatology, Department of Pediatrics, Children’s Hospital of Philadelphia and University of Pennsylvania Perelman School of Medicine, Philadelphia, PA, USA
Correspondence: Xiaohui Wang; Huayan Zhang, Email [email protected]; [email protected]
Background: Bronchopulmonary dysplasia (BPD) has become a major cause of morbidity and mortality in preterm infants worldwide, yet its pathogenesis and underlying mechanisms remain poorly understood. The present study sought to explore microRNA-mRNA regulatory networks and immune cells involvement in BPD through a combination of bioinformatic analysis and experimental validation.
Methods: MicroRNA and mRNA microarray datasets were obtained from the Gene Expression Omnibus (GEO) database. Differentially expressed microRNAs (DEMs) were identified in BPD patients compared to control subjects, and their target genes were predicted using miRWalk, miRNet, miRDB, and TargetScan databases. Subsequently, protein-protein interaction (PPI) and functional enrichment analyses were conducted on the target genes. 30 hub genes were screened using the Cytohubba plugin of the Cytoscape software. Additionally, mRNA microarray data was utilized to validate the expression of hub genes and to perform immune infiltration analysis. Finally, real-time PCR (RT-PCR), immunohistochemistry (IHC), and flow cytometry were conducted using a mouse model of BPD to confirm the bioinformatics findings.
Results: Two DEMs (miR-15b-5p and miR-20a-5p) targeting genes primarily involved in the regulation of cell cycle phase transition, ubiquitin ligase complex, protein serine/threonine kinase activity, and MAPK signaling pathway were identified. APP and four autophagy-related genes (DLC1, PARP1, NLRC4, and NRG1) were differentially expressed in the mRNA microarray dataset. Analysis of immune infiltration revealed significant differences in levels of neutrophils and naive B cells between BPD patients and control subjects. RT-PCR and IHC confirmed reduced expression of APP in a mouse model of BPD. Although the proportion of total neutrophils did not change appreciably, the activation of neutrophils, marked by loss of CD62L, was significantly increased in BPD mice.
Conclusion: Downregulation of APP mediated by miR-15b-5p and miR-20a-5p may be associated with the development of BPD. Additionally, increased CD62L− neutrophil subset might be important for the immune-mediated injury in BPD.
Keywords: bronchopulmonary dysplasia, CD62L− neutrophil, immune infiltration analysis, miRNA-mRNA regulatory circuits
Introduction
Bronchopulmonary dysplasia (BPD), characterized by impaired alveolarization and dysregulated vascularization in the lung, is an important cause of morbidity and mortality in preterm infants worldwide.1 The younger the gestational age and the lower the birth weight, the higher the incidence and severity of BPD, with infants born at less than 1, 250 g accounting for 97% of BPD cases.2 Advances in neonatal intensive care have improved the survival of preterm infants, but not the incidence of BPD. BPD survivors often have cardiopulmonary and neurological sequelae, resulting in heavy medical care and financial burdens on families and society.3 Unfortunately, the etiology and pathogenesis of BPD are still ambiguous so far.4,5
In the past decades, immune reactions and inflammation have been topics of interest to investigators on the mechanisms of BPD.6 Besides hyperoxia exposure, antenatal and postnatal inflammation induced by injection of LPS could also cause BPD in animal models.7,8 Immune cell-derived inflammatory cytokines, including IL-1, IL-6, TNF-α, were reported to be positively correlated with BPD severity.9–11 Nevertheless, although excessive inflammatory responses were recognized to be key risk factors for BPD, which immune cell types played important roles over the course of BPD progression remained obscure. Numerous studies have reported that microRNAs (miRNAs) could affect BPD through multiple mechanisms, involving regulation of angiogenesis, extracellular matrix, and inflammation.12–14 However, whether miRNA-mRNA regulatory circuits could regulate specific immune cells and thus affect BPD processes is not yet clear.
In this study, we compared miRNA and mRNA expression between BPD patients and controls through datasets obtained from the Gene Expression Omnibus (GEO). Then, miRNA-mRNA regulatory networks were constructed, and enrichment analysis of target genes was performed. Furthermore, we screened hub genes by protein-protein interaction (PPI) network analysis and inferred the proportion of 22 immune cell types in BPD using the CIBERSORT algorithm. Finally, we conducted real-time PCR (RT-PCR), immunohistochemistry (IHC), and flow cytometry to validate our results of bioinformatic analyses in BPD mice. Together, using bioinformatic techniques followed by confirmation in an experimental mouse model, this study aimed to investigate miRNA-mRNA regulatory circuits and immune signatures associated with BPD, which might improve current understanding of the mechanisms underlying BPD and provide clues to potential therapeutic targets for BPD.
Materials and Methods
Gene Expression Data Acquisition
The miRNA expression dataset (GSE108755) and mRNA expression dataset (GSE108754) were acquired from the GEO database (http://www.ncbi.nlm.nih.gov/geo). Each of the two datasets included blood samples from 5 BPD patients born at 28 weeks’ gestational age and 6 non-BPD age-matched controls. A flow diagram demonstrating the overall design of the study was shown in Figure 1.
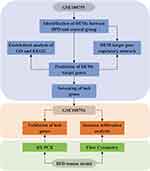 |
Figure 1 Flow chart of the whole study. |
Identification of Differentially Expressed miRNAs (DEMs)
The gene expression matrix from GSE108755 and its corresponding platform annotation file were downloaded from the GEO database. The matrix was log2 transformed and quantile normalized using the “Normalize Between Arrays” function of “limma” package under R (version 4.2.1).15 Next, differential expression analysis between the BPD group and controls was performed using the limma algorithm. The DEMs were screened at a threshold of p-value of 0.05 and |log2 fold change (logFC)| of 1.5. The “ggplot2” package was used to draw the heatmap.16
Construction of DEMs-Target Genes Interaction Network
The potential target genes of DEMs were predicted through the miRWalk, miRNet, miRDB, and TargetScan databases.17–20 Concordant target genes of the four databases were identified by drawing a Venn diagram using an online tool (http://bioinformatics.psb.ugent.be/webtools/Venn/). The DEMs-target genes interaction network was visualized using Cytoscape software (version 3.8.2).21
Functional Enrichment Analysis
Gene ontology (GO) and kyoto encyclopedia of genes and genomes (KEGG) analysis were performed using the DAVID functional annotation tool (version 6.8). GO terms include 3 categories: biological process (BP), cellular component (CC), and molecular function (MF). GO terms and KEGG pathways with a p-value < 0.05 and q value < 0.2 were considered to be significantly enriched. Visualization of the results of enrichment analysis was conducted using the “ggplot2” package in R.
PPI Network and Identification of Hub Genes
The PPI network of target genes was established using the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database (http://string-db.org/) with default parameters.22 Then, the interaction file containing the source and target nodes was downloaded and imported into Cytoscape software. CytoHubba, a plugin of Cytoscape, was used to rank the nodes. The top 30 nodes in the PPI network were selected as hub genes based on the maximal clique centrality (MCC) method, which was recognized to have a better performance on precise predictions of essential proteins.23
Prediction of Upstream Transcription Factors of Hub Genes
The ChEA3 database, which contains 6 primary reference gene set libraries created from multiple resources, was used to predict the potential upstream transcription factors of hub genes.24 In this study, the ENCODE library was selected.
Autophagy-Related Genes (ARGs)
To investigate the involvement of autophagy in the pathogenesis of BPD, a total of 222 genes related to autophagy were obtained from the Human Autophagy Database (http://www.autophagy.lu/index.html).
Validation of Hub Genes and ARGs
GSE108754, available in the GEO database, was used to validate the expression of hub genes. Differential gene expression analysis was performed using the “limma” package. Genes with p-value < 0.05 and |logFC| >1 were considered as differentially expressed genes (DEGs). Valid hub genes need to satisfy both of the following criteria:1) the direction of hub genes should be opposite to their corresponding DEMs; 2) DEGs. The intersections of DEGs and ARGs were also selected.
Immune Infiltration Analysis
Immune infiltration analysis of 22 types of immune cells was conducted using the CIBERSORT algorithm in R, and the results were visualized using the “ggplot2” package.25
BPD Model and Tissue Collection
Neonatal C57BL/6J mice in this study were purchased from Guangzhou Zhiyuan Biotechnology. Mice were randomly assigned to the room air (RA) group (exposure to room air, n = 15) and hyperoxia group (exposure to 85% oxygen, n = 16) within the first 24 hours after birth. Dams were exchanged between hyperoxia and RA groups every 24 hours. Lung tissues of mice on day 7 after birth were collected for flow cytometry (n = 11). Other mice were sacrificed 14 days after birth for experiments except flow cytometry (n = 20).
RNA Extraction and RT-PCR
Total RNA was isolated from mice lungs with the RNA-easy Isolation Reagent (Vazyme, China). The total RNA was reverse transcribed using HiScript III RT SuperMix for qPCR (+ gDNA wiper) (Vazyme, China) analysis, ChamQ Universal SYBR qPCR Master Mix (Vazyme, China). RT-PCR was performed using QuantStudio 6 Flex real-time PCR system (Applied Biosystems, USA). Primer sequences are displayed in Table 1.
 |
Table 1 Primers Used for RT-PCR of Mouse Genes |
IHC
The lung paraffin sections were dewaxed to water, and then antigen retrieval was performed with pH 9.0 Antigen Retrieval Solution. After blocking endogenous peroxidase with 3% H2O2, the sections were blocked with 5% fetal bovine serum and incubated with the primary rabbit monoclonal anti-amyloid precursor protein antibody (ab32136, Abcam) at 4°C overnight. Sections were washed with PBS, then incubated with HRP-conjugated goat anti-rabbit IgG (ab6721, Abcam) for 1 hour at room temperature. After washing sections, the DAB substrate was added to the sections until color developed. Finally, the sections were examined using a Leica microscope and photographed.
Flow Cytometry
On day 7, the lungs were isolated from mice, grounded, and subsequently passed through a 70μm cell strainer. Ammoniumchloride-potassium lysing buffer was used for erythrocyte lysis. Multiparameter assessments were performed using BD FACSAria II (BD Biosciences) and data was analyzed with Flowjo software (version 10.6). After staining Zombie Aqua™ (Biolegend) to exclude dead cells, single-cell suspensions were incubated with the fluorochrome-conjugated antibodies in PBS containing 2% fetal bovine serum and then washed twice before detection. Antibodies specific to mouse used included CD16/32 (BD, 553,141), CD45-APC-Cy7 (Biolegend, 103,116), CD11b-AF700 (Biolegend, 101,222), Ly6G-PE (Biolegend, 127,608), CD62L-BV650 (Biolegend, 104,453), CD3-BV711 (Biolegend, 100,241), CD19-BV711 (Biolegend, 115,555), NK1.1-Pacific Blue (Biolegend, 109,816).
Statistical Analysis
Statistical analysis was performed using R (version 4.2.1) or GraphPad Prism (version 9). Data were expressed as mean ± SD. Comparisons of means between two groups were done with Student’s t-test. For correlation analysis, Spearman’s method was used. p-value < 0.05 was considered statistically significant.
Results
Identification of DEMs
The GSE108755 dataset was downloaded from the GEO database. After log2 transformation and normalization, differential expression analysis was performed using the “limma” package in R. The results were shown as a heatmap in Figure 2A. Two miRNAs (miR-15b-5p and miR-20a-5p) were finally identified to be DEMs.
Construction of DEMs-Target Genes Network
The potential target genes of DEMs were predicted using the miRWalk, miRNet, miRDB, and TargetScan databases, and 157 target genes were eventually screened (Figure 2B). The DEMs-target genes network was constructed using Cytoscape software (Figure 2C).
Functional Enrichment Analysis of Target Genes
To gain further insight into functions of the 157 target genes of DEMs, GO terms enrichment and KEGG pathway analysis were performed. BP analysis revealed that DEMs target genes were mainly enriched in myeloid cell differentiation, regulation of cell cycle phase transition, and macroautophagy (Figure 3A). CC analysis demonstrated that DEMs target genes were enriched in RNA polymerase II transcription regulator complex (Figure 3A). As for MF, DEMs target genes were enriched in protein serine/threonine kinase activity, protein-macromolecule adaptor activity, and SH3 domain binding (Figure 3A). KEGG pathway analysis indicated that target genes were involved in cellular senescence, cell cycle, MAPK signaling pathway, TGF-beta signaling pathway, VEGF signaling pathway, etc (Figure 3B).
Construction of PPI Network and Screening of Hub Genes
A PPI network was established with the 157 target genes using the STRING database. Then, the interaction file was downloaded and imported into Cytoscape and processed with the cytoHubba plugin to identify hub genes. The top 30 hub genes are shown in Figure 4A. For hsa-miR-20a-5p, the hub genes were WEE1, E2F2, E2F3, MAPK1, CREB1, RBL2, AGO1, VEGFA, TNRC6B, USP28, APP, E2F5, CRK, SMAD5, YOD1, BCL2, OTUD4, CLOCK, REST and RGMB. For hsa-miR-15b-5p, the hub genes were CCND3, WEE1, E2F3, CHEK1, MAPK1, VEGFA, TNRC6B, BTRC, AKT3, APP, SMURF1, CRK, CLSPN, ACVR2A, PSMD7, BCL2, PHC3 and CDC27.
Prediction of Upstream Transcription Factors of Hub Genes
The upstream transcription factors of the top 30 hub genes, including ZNF384, BHLHE40, E2F4, GATA2, CTCFL, and CEBPB, were predicted using ChEA3 (Figure 4B).
Validation of Hub Genes and ARGs
The expression of hub genes was further verified using the GSE108754 dataset. However, only amyloid beta precursor protein (APP) was significantly downregulated in the BPD group compared with controls (Figure 5). The expression of CHEK1, BCL2, CCND3, E2F3, CLSPN, PSMD7, VEGFA, SMAD5, CLOCK, and AKT3 were consistently decreased but did not reach statistical significance (p-value > 0.05 or |logFC| < 1). ARGs expression levels were also assessed. Using the cutoff value of p-value < 0.05 and |logFC| > 1, two ARGs were found to be upregulated (DLC1 and PARP1) and two downregulated (NLRC4 and NRG1) (Table 2).
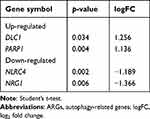 |
Table 2 Differentially Expressed ARGs |
 |
Figure 5 Validation of hub genes in the GSE108754 dataset. Student’s t-test. *p < 0.05. ns not significant. |
Immune Cell Infiltration Analysis
The relative proportions of 22 types of immune cells in BPD and normal samples were inferred using the CIBERSORT algorithm in R. Figure 6A showed the composition of immune cells in each sample of the GSE108754 dataset. The correlation heatmap of immune cells demonstrated that there was a strong positive correlation between resting dendritic cells and Tregs (r > 0.8). Resting NK cells and activated CD4+ memory T cells also had a strong positive correlation, while monocytes and CD8+ T cells were strongly negatively correlated (r < −0.8). Neutrophils were positively correlated with resting CD4+ memory T cells and negatively correlated with naïve B cells (Figure 6B). Box plots showed that naïve B cells were significantly increased in the BPD group, while neutrophils were significantly reduced (Figure 6C). Interestingly, the expression of APP was found to be negatively correlated with neutrophils (Figure 6B).
Validation of Hub Genes in Mouse Models of BPD
Consistent with the phenotypes of human BPD, hyperoxia-induced BPD mice exhibited disruption of alveolar growth and enlargement of alveolar spaces (Figure 7A), with decreased radial alveolar count (RAC) and increased mean linear intercept (MLI) (Figure 7B). The expression of genes previously identified was verified using RT-PCR in BPD mice. The expression level of App was significantly reduced in the lungs of BPD mice compared with the RA group. However, contrary to our expectation, Nrg1 expression was elevated (p > 0.05). Parp1, Nlrc4, and Dlc1 showed trends consistent with previous bioinformatic analyses, but the differences did not reach statistical significance (p > 0.05) (Figure 7C). IHC staining further confirmed the downregulation of APP levels in BPD mice (Figure 7D).
CD62L+ Neutrophils Significantly Decreased in BPD Mice
Through flow cytometry, we found that the proportion of total neutrophils did not differ between the RA and BPD groups (Figure 7E). However, the population of CD62L+ neutrophil subset (p < 0.01) was dramatically decreased, whereas the population of CD62L− neutrophil subset (p < 0.01) was significantly increased in BPD mice compared with controls (Figure 7F and G). These findings suggest that in response to hyperoxia, the activation status in neutrophils altered in BPD mice, suggesting a potential role of neutrophils in hyperoxia-induced injury.
Discussion
Despite rapid advances in medical technology, the incidence of BPD remains high, and effective therapies are lacking, largely due to limited understanding of its pathogenesis. The mechanisms underlying BPD are complex, involving ferroptosis, IL-6, Wnt signaling, etc.9,26,27
This study aimed to identify the miRNA-mRNA regulatory circuits that are most likely related to BPD and explore the roles of immune cells in BPD pathogenesis. Two miRNAs (miR-15b-5p and miR-20a-5p), both upregulated, were identified as DEMs. Among them, miR-15b-5p was reported to regulate the proliferation and apoptosis of human vascular smooth muscle cells.28 miR-20a-5p was found to inhibit autophagy in ovarian cancer.29 Notably, the role that these two miRNAs play in BPD is still unclear.
GO enrichment analysis showed that DEMs target genes were mainly involved in myeloid cell differentiation, regulation of cell cycle phase transition, macroautophagy, ubiquitin ligase-substrate adaptor activity, and protein serine/threonine kinase activity. KEGG analysis showed that target genes were mainly enriched in the cell cycle, MAPK signaling pathway, TGF-β signaling pathway, and VEGF signaling pathway. Through PPI network analysis, 30 hub genes were screened. However, among these hub genes, only APP was significantly decreased in the mRNA expression dataset (p-value < 0.05 and |logFC| > 1). Significant downregulation of APP was also confirmed by RT-PCR and IHC in the mouse model of BPD. APP is a highly pleiotropic protein involved in numerous cellular functions.30 APP misprocessing and deposition of pathogenic fragments could cause neuron and synapse loss, which is the most recognized hypothesis in the pathogenesis of Alzheimer’s disease.31 However, according to the Bio Gene Portal System (BioGPS, http://biogps.org/) database, apart from the brain, APP is widely expressed in many tissues, including the bronchus and lung. APP was reported to regulate global protein synthesis in a variety of human dividing cells.32 Depletion of APP could cause cell size abnormalities and death through G0 arrest.33 To date, the exact function of APP in the lung has not been reported. Previous studies reported that APP played an important protective function in cerebral vascular endothelium by maintaining endothelial nitric oxide synthase (eNOS) expression and mediating reactivity to vascular endothelial growth factor (VEGF).34,35 Aberrant pulmonary alveolarization and dysregulated vascularization are key features of BPD, and impaired VEGF and eNOS signaling have been proved to be related to BPD progression.36,37 Herein, we speculated that miR-15b-5p/miR-20a-5p-APP might promote BPD via impairing cell cycle and VEGF/eNOS signaling. Dysregulation of autophagy has been reported to be associated with various lung diseases.38,39 However, the role that autophagy plays in BPD pathogenesis is not clear. We identified 4 different ARGs (DLC1, PARP1, NLRC4 and NRG1) in BPD and verified them via RT-PCR, but none of the differences reached statistical significance.
Exaggerated immune activation and inflammation are key risk factors for BPD.40 Continued inflammation recruits immune cells, which in turn secrete more pro-inflammatory cytokines, ultimately aggravating the inflammation and causing lung injury. Neutrophils are the earliest immune cells to infiltrate the damaged tissue after injury and have been proven to be associated with chronic lung diseases.41 However, little research has been conducted regarding changes of circulating or lung neutrophils in BPD. In this study, we estimated the composition of immune cells in BPD patients and controls using the CIBERSORT algorithm. We found that neutrophils were reduced in the peripheral blood of BPD patients, which was contrary to some previous reports.42–44 Reasons for this inconsistency are not clear, partly due to different gestational ages and postnatal days as the number of neutrophils in the peripheral blood change drastically within the first week after birth. Besides, it was reported that neutrophils remained in the pulmonary vascular bed during acute lung injury, leading to a reduction of peripheral blood neutrophil counts.45 Interestingly, APP levels were strongly negatively correlated with the proportion of neutrophils.
In the present study, we found that lung neutrophils in newborn mice could be divided into two subgroups: CD62L+ neutrophils and CD62L− neutrophils, and the proportion of CD62L− neutrophil subset was dramatically increased in the lungs of BPD mice. As CD62L shedding is a sign of neutrophil activation, the decreased expression of surface CD62L suggests that pulmonary neutrophils are more activated in BPD mice.46,47 It was reported that alveolar neutrophils with reduced CD62L expression display delayed apoptosis and enhanced respiratory burst in acute respiratory distress syndrome.48 The Wnt pathway is critical both during embryonic lung development and in BPD.26 Recently, it was reported that neutrophil extracellular traps (NETs) could promote BPD progression via the Wnt/β-catenin pathway.49 As NETs are released by activated neutrophils, it is possible that CD62L− neutrophil subset promotes BPD progression via NETs. Further experiments are required to test this hypothesis.
In all, based on bioinformatic analyses, we constructed miRNA-mRNA regulatory networks, which enabled a more comprehensive understanding of genes and interactions related to BPD. In addition, we estimated changes of immune cell proportions in BPD patients applying CIBERSORT. Finally, we performed RT-PCR, IHC and flow cytometry in BPD mice to confirm our findings. However, our study also has some limitations. First, our study relied on public datasets with insufficient sample size and incomplete clinical information, which might bias the results and limit the generalizability of the study findings. Second, the specific role and mechanism of APP in BPD pathogenesis remains unclear, further experiments, particularly single-cell RNA sequencing, are warranted. Besides, the function of CD62L− neutrophil subset and its role in BPD pathogenesis deserve particular attention in future research.
Conclusion
The results of this study indicated that the downregulation of APP mediated by miR-15b-5p and miR-20a-5p might be associated with BPD progression. Besides, increased CD62L− neutrophil subset may promote BPD progression via excessive inflammatory responses. Our study might help to understand the pathogenesis of BPD and provide potential therapeutic targets for BPD.
Data Sharing Statement
The datasets we utilized in this study are available in the GEO database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc = ) under accession number GSE108754 and GSE108755.
Ethics
The studies involving human data from the GEO database were reviewed and approved by the Ethics Committee of Guangzhou Women and Children’s Medical Center (No. 050A01, 1 April 2023). All animal experiments conformed to the Guidelines for Care and Use of Laboratory Animals and were approved by the Ethics Committee of Guangzhou Women and Children’s Medical Center (No. 307B00, 10 May 2022).
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis, and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was supported by the Science and Technology Program of Guangzhou, China (Grant No. 2023A04J1882), National Natural Science Foundation of China (Grant No. 82201946), and Guangzhou Municipal Science and Technology Bureau (Grant No. SL2022A03J01156).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Higgins RD, Jobe AH, Koso-Thomas M, et al. Bronchopulmonary dysplasia: executive summary of a workshop. J Pediatr. 2018;197:300–308. doi:10.1016/j.jpeds.2018.01.043
2. Walsh MC, Szefler S, Davis J, et al. Summary proceedings from the bronchopulmonary dysplasia group. Pediatrics. 2006;117(3):S52–6. doi:10.1542/peds.2005-0620I
3. Islam JY, Keller RL, Aschner JL, et al. Understanding the short- and long-term respiratory outcomes of prematurity and bronchopulmonary dysplasia. Am J Respir Crit Care Med. 2015;192(2):134–156. doi:10.1164/rccm.201412-2142PP
4. Principi N, Di Pietro GM, Esposito S. Bronchopulmonary dysplasia: clinical aspects and preventive and therapeutic strategies. J Transl Med. 2018;16(1):36. doi:10.1186/s12967-018-1417-7
5. Thomas JM, Sudhadevi T, Basa P, et al. The role of sphingolipid signaling in oxidative lung injury and pathogenesis of bronchopulmonary dysplasia. Int J Mol Sci. 2022;23(3). doi:10.3390/ijms23031254
6. Zaramella P, Munari F, Stocchero M, et al. Innate immunity ascertained from blood and tracheal aspirates of preterm newborn provides new clues for assessing bronchopulmonary dysplasia. PLoS One. 2019;14(9):e0221206. doi:10.1371/journal.pone.0221206
7. Berger J, Bhandari V. Animal models of bronchopulmonary dysplasia. The term mouse models. Am J Physiol Lung Cell Mol Physiol. 2014;307(12):L936–47. doi:10.1152/ajplung.00159.2014
8. Xu J, Mao X, Jin R, et al. Neutrophil extracellular traps degrade fibronectin in a rat model of bronchopulmonary dysplasia induced by perinatal exposure to lipopolysaccharide. J Cell Mol Med. 2020;24(24):14645–14649. doi:10.1111/jcmm.15842
9. Windhorst AC, Heydarian M, Schwarz M, et al. Monocyte signature as a predictor of chronic lung disease in the preterm infant. Front Immunol. 2023;14:1112608. doi:10.3389/fimmu.2023.1112608
10. Eldredge LC, Creasy RS, Presnell S, et al. Infants with evolving bronchopulmonary dysplasia demonstrate monocyte-specific expression of IL-1 in tracheal aspirates. Am J Physiol Lung Cell Mol Physiol. 2019;317(1):L49–l56. doi:10.1152/ajplung.00060.2019
11. Sahoo D, Zaramela LS, Hernandez GE, et al. Transcriptional profiling of lung macrophages identifies a predictive signature for inflammatory lung disease in preterm infants. Commun Biol. 2020;3(1):259. doi:10.1038/s42003-020-0985-2
12. Hu Y, Xie L, Yu J, et al. Inhibition of microRNA-29a alleviates hyperoxia-induced bronchopulmonary dysplasia in neonatal mice via upregulation of GAB1. Mol Med. 2019;26(1):3. doi:10.1186/s10020-019-0127-9
13. Syed M, Das P, Pawar A, et al. Hyperoxia causes miR-34a-mediated injury via angiopoietin-1 in neonatal lungs. Nat Commun. 2017;8(1):1173. doi:10.1038/s41467-017-01349-y
14. Durrani-Kolarik S, Pool CA, Gray A, et al. miR-29b supplementation decreases expression of matrix proteins and improves alveolarization in mice exposed to maternal inflammation and neonatal hyperoxia. Am J Physiol Lung Cell Mol Physiol. 2017;313(2):L339–l49. doi:10.1152/ajplung.00273.2016
15. Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi:10.1093/nar/gkv007
16. Ito K, Murphy D. Application of ggplot2 to pharmacometric graphics. CPT Pharmacometrics Syst Pharmacol. 2013;2(10):e79. doi:10.1038/psp.2013.56
17. Chen Y, Wang X. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020;48(D1):D127–d31. doi:10.1093/nar/gkz757
18. Fan Y, Siklenka K, Arora SK, et al. miRNet - dissecting miRNA-target interactions and functional associations through network-based visual analysis. Nucleic Acids Res. 2016;44(W1):W135–41. doi:10.1093/nar/gkw288
19. Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi:10.1016/j.cell.2004.12.035
20. Sticht C, De La Torre C, Parveen A, et al. miRWalk: an online resource for prediction of microRNA binding sites. PLoS One. 2018;13(10):e0206239. doi:10.1371/journal.pone.0206239
21. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi:10.1101/gr.1239303
22. Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–d13. doi:10.1093/nar/gky1131
23. Chin CH, Chen SH, Wu HH, et al. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 2014;8(4):S11. doi:10.1186/1752-0509-8-s4-s11
24. Keenan AB, Torre D, Lachmann A, et al. ChEA3: transcription factor enrichment analysis by orthogonal omics integration. Nucleic Acids Res. 2019;47(W1):W212–w24. doi:10.1093/nar/gkz446
25. Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–457. doi:10.1038/nmeth.3337
26. Lingappan K, Savani RC. The wnt signaling pathway and the development of bronchopulmonary dysplasia. Am J Respir Crit Care Med. 2020;201(10):1174–1176. doi:10.1164/rccm.202002-0277ED
27. Hu Z, Liu C, Mao Y, et al. Integration of transcriptomics reveals ferroptosis-related signatures and immune cell infiltration in bronchopulmonary dysplasia. Heliyon. 2023;9(10):e21093. doi:10.1016/j.heliyon.2023.e21093
28. Song N, Wang W, Wang Y, et al. Hydrogen sulfide of air induces macrophage extracellular traps to aggravate inflammatory injury via the regulation of miR-15b-5p on MAPK and insulin signals in trachea of chickens. Sci Total Environ. 2021;771:145407. doi:10.1016/j.scitotenv.2021.145407
29. Li H, Lei Y, Li S, et al. MicroRNA-20a-5p inhibits the autophagy and cisplatin resistance in ovarian cancer via regulating DNMT3B-mediated DNA methylation of RBP1. Reprod Toxicol. 2022;109:93–100. doi:10.1016/j.reprotox.2021.12.011
30. Müller UC, Zheng H. Physiological functions of APP family proteins. Cold Spring Harb Perspect Med. 2012;2(2):a006288. doi:10.1101/cshperspect.a006288
31. Sharma C, Kim S, Nam Y, et al. Mitochondrial dysfunction as a driver of cognitive impairment in alzheimer’s disease. Int J Mol Sci. 2021;22(9):4850. doi:10.3390/ijms22094850
32. Sobol A, Galluzzo P, Liang S, et al. Amyloid precursor protein (APP) affects global protein synthesis in dividing human cells. J Cell Physiol. 2015;230(5):1064–1074. doi:10.1002/jcp.24835
33. Sobol A, Galluzzo P, Weber MJ, et al. Depletion of amyloid precursor protein (APP) causes G0 arrest in non-small cell lung cancer (NSCLC) cells. J Cell Physiol. 2015;230(6):1332–1341. doi:10.1002/jcp.24875
34. d’Uscio LV, He T, Santhanam AV, et al. Endothelium-specific amyloid precursor protein deficiency causes endothelial dysfunction in cerebral arteries. J Cereb Blood Flow Metab. 2018;38(10):1715–1726. doi:10.1177/0271678x17735418
35. Ristori E, Cicaloni V, Salvini L, et al. Amyloid-β precursor protein APP down-regulation alters actin cytoskeleton-interacting proteins in endothelial cells. Cells. 2020;9(11):2506. doi:10.3390/cells9112506
36. Abman SH. Bronchopulmonary dysplasia “a vascular hypothesis”. Am J Respir Crit Care Med. 2001;164(10 Pt 1):1755–1756. doi:10.1164/ajrccm.164.10.2109111c
37. Seedorf G, Metoxen AJ, Rock R, et al. Hepatocyte growth factor as a downstream mediator of vascular endothelial growth factor-dependent preservation of growth in the developing lung. Am J Physiol Lung Cell Mol Physiol. 2016;310(11):L1098–110. doi:10.1152/ajplung.00423.2015
38. Xia F, Deng C, Jiang Y, et al. IL4 (interleukin 4) induces autophagy in B cells leading to exacerbated asthma. Autophagy. 2018;14(3):450–464. doi:10.1080/15548627.2017.1421884
39. Zhang F, Ma H, Wang ZL, et al. The PI3K/AKT/mTOR pathway regulates autophagy to induce apoptosis of alveolar epithelial cells in chronic obstructive pulmonary disease caused by PM2.5 particulate matter. J Int Med Res. 2020;48(7):300060520927919. doi:10.1177/0300060520927919
40. Salimi U, Dummula K, Tucker MH, et al. Postnatal sepsis and bronchopulmonary dysplasia in premature infants: mechanistic insights into “New BPD”. Am J Respir Cell Mol Biol. 2022;66(2):137–145. doi:10.1165/rcmb.2021-0353PS
41. Voynow JA, Shinbashi M. Neutrophil elastase and chronic lung disease. Biomolecules. 2021;11(8):1065. doi:10.3390/biom11081065
42. Li X, Wang Q, Luo T, et al. Decreased neutrophil levels in bronchopulmonary dysplasia infants. Pediatr Neonatol. 2020;61(6):637–644. doi:10.1016/j.pedneo.2020.08.013
43. Palta M, Sadek-Badawi M, Carlton DP. Association of BPD and IVH with early neutrophil and white counts in VLBW neonates with gestational age <32 weeks. J Perinatol. 2008;28(9):604–610. doi:10.1038/jp.2008.65
44. Sun Y, Chen C, Zhang X, et al. High neutrophil-to-lymphocyte ratio is an early predictor of bronchopulmonary dysplasia. Front Pediatr. 2019;7:464. doi:10.3389/fped.2019.00464
45. Potera RM, Cao M, Jordan LF, et al. Alveolar macrophage chemokine secretion mediates neutrophilic lung injury in Nox2-deficient mice. Inflammation. 2019;42(1):185–198. doi:10.1007/s10753-018-0883-7
46. Geng S, Zhang Y, Lee C, et al. Novel reprogramming of neutrophils modulates inflammation resolution during atherosclerosis. Sci Adv. 2019;5(2):eaav2309. doi:10.1126/sciadv.aav2309
47. Tak T, Wijten P, Heeres M, et al. Human CD62L(dim) neutrophils identified as a separate subset by proteome profiling and in vivo pulse-chase labeling. Blood. 2017;129(26):3476–3485. doi:10.1182/blood-2016-07-727669
48. Juss JK, House D, Amour A, et al. Acute respiratory distress syndrome neutrophils have a distinct phenotype and are resistant to phosphoinositide 3-kinase inhibition. Am J Respir Crit Care Med. 2016;194(8):961–973. doi:10.1164/rccm.201509-1818OC
49. Sun L, Zhang M, Jiang J, et al. Neutrophil extracellular traps promote bronchopulmonary dysplasia-like injury in neonatal mice via the WNT/β-catenin pathway. Front Cell Infect Microbiol. 2023;13:1126516. doi:10.3389/fcimb.2023.1126516
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.





