Back to Journals » Journal of Asthma and Allergy » Volume 16
Interleukin-4 (C590T) Gene Polymorphism in Association with Asthma Severity
Authors Al-Ahmad M, Ali A , Haider MZ
Received 11 July 2023
Accepted for publication 10 November 2023
Published 21 November 2023 Volume 2023:16 Pages 1269—1278
DOI https://doi.org/10.2147/JAA.S429981
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Amrita Dosanjh
Mona Al-Ahmad,1,2,* Asmaa Ali,2– 4,* Mohammad Z Haider5
1Department of Microbiology, College of Medicine, Kuwait University, Kuwait City, Kuwait; 2Department of Allergy, Al-Rashed Allergy Center, Ministry of Health, Kuwait City, Kuwait; 3Department of Laboratory Medicine, School of Medicine, Jiangsu University, Zhenjiang, People’s Republic of China; 4Department of Pulmonary Medicine, Abbassia Chest Hospital, Ministry of Health, Cairo, Egypt; 5Department of Pediatrics, College of Medicine, Kuwait University, Kuwait City, Kuwait
*These authors contributed equally to this work
Correspondence: Mona Al-Ahmad, Department of Microbiology, College of Medicine, Kuwait University, Kuwait City, Kuwait, Tel +965-24636515, Fax +965-25332719, Email [email protected]
Background: A significant link between T allele of the IL-4 (C590T) gene and developing asthma in some populations was reported. However, no study discussed the link between IL-4 (C590T) gene polymorphism and asthma severity groups (mild and severe). This study investigated the link between IL-4 gene variation and asthma severity.
Methods: The study included 215 asthmatic patients, of which 102 had mild asthma, and 126 participants were healthy controls. A previously published polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) method was used to identify various IL-4 (C590T) gene polymorphism genotypes.
Results: The T allele frequency was higher in mild asthma (p=0.002) but not in severe asthma (p=0.12) compared to controls. In mild asthma, the CT genotype and (CT+TT versus CC) increased the likelihood of asthma threefold (p< 0.001, 0.001). However, no significant association with severe asthma was found in either genetic model. Stratification analysis showed that the C allele and CC genotype increased the risk of severe asthma (p=0.01). The recessive genetic model indicated a decrease in the risk of severe asthma (OR=0.5, p=0.01) in the non-adjusted regression analysis. Adjusting for age, sex, and other risk factors revealed that the IL-4 gene polymorphism did not influence the risk of severe asthma (OR=0.92, p=0.80); however, being an elderly female with a history of childhood-onset disease and associated nasal polyp (NP) increased the likelihood of severe asthma, OR=1.08, 2.01, 2.36, 8.42; p< 0.001, 0.05, 0.05, < 0.001, respectively.
Conclusion: The T allele and CT genotype in the co-dominant genetic model and the (CT+TT) genotype in the recessive model were found to have a higher likelihood of developing mild asthma but not severe asthma; severe asthma was found to be higher in elderly females with a history of childhood-onset disease and associated nasal polyps.
Keywords: IL-4 (C590T) gene polymorphism, mild and severe asthma, nasal polyp
Introduction
Asthma is a common chronic inflammatory disease affecting more than 330 million people worldwide and could develop at any age;1 however, the first asthma symptoms mostly appear in the childhood.2 Episodes of bronchial hyper-responsiveness and reversible airway obstruction are the characteristic hallmarks of asthma.3 The main causes of asthma are not known; however, several factors trigger the pathogenesis cascade of asthma, which include environmental triggers such as pollutants, tobacco smoking, allergens, viral and bacterial infection, and genetic variations.4,5 The interaction between all these underlying risk factors increases the possibility of asthma presentation and even could determine the disease severity and control.5 The pathogenesis of asthma is complex, and several immune cells and biomarkers are involved including T-lymphocyte, mast cells and eosinophils, and cytokines such as IL-4, IL-5, and IL-13 plus the immunoglobulin-E (IgE).6,7
Interleukin-4 (IL-4) has been shown to play a crucial role in asthma pathogenesis; it influences the isotype switch to the ε immunoglobulin class, which leads to the production and secretion of immunoglobulin E (IgE) by B lymphocytes.8 High levels of IgE are a hallmark of allergic diseases, and IL-4 is considered a vital mediator in this process. Additionally, this cytokine is involved in developing airway obstruction through the induction of mucin gene expression, leading to exaggerated mucus production, and contributing to the characteristic airway hyperresponsiveness seen in asthma.9 Furthermore, IL-4 also increases the expression of eotaxin and other inflammatory cytokines from fibroblasts, which contribute to airway inflammation and lung remodeling in chronic asthma.9,10
The IL-4 gene is located on chromosome 5q31-33 and codes for a protein that binds to the IL-4 receptor.11 It is highly polymorphic, and divergences in its sequence have been associated with susceptibility to several immune-mediated diseases, including asthma, allergies, and autoimmune disorders.11–13 Five putative variants have been reported in the promoter region of IL-4 gene, of which four are rare.14 The variant – C590T in the IL-4 gene’s promoter region has been associated with increased IgE levels in American15 and Japanese populations.16 In-vitro studies suggest that this variant may affect IL-4 activity by altering the binding affinity to nuclear transcription factors and electrophoretic mobility.15 However, there is no direct evidence linking this variant to cellular IgE synthesis, and the original association has not been consistently reproduced. Two additional variants have been identified but are not associated with atopy or asthma.11 The C590T polymorphism in the IL-4 gene has been extensively studied concerning asthma, and evidence suggests that it may contribute to the development of the disease, as well as its response to treatment.17
However, to date, there is no study reporting a link between IL-4 (C590T) gene polymorphism and severe asthma, and whether there is any difference in its association with mild and/or severe asthma along with other risk factors. This study aimed to investigate whether there is a significant difference in the association of IL-4 gene C590T polymorphism between mild and severe asthma.
Methods
Patients and Study Design
This case-control study was conducted at Al-Rashed Allergy Center, Kuwait, from October 2022 to March 2023. The study enrolled 215 patients with confirmed bronchial asthma based on clinical diagnosis and reversibility of FEV1%.18 Additionally, 126 healthy controls with no history of asthma, atopy, or other allergic conditions were included in the study. All participants were Kuwaiti in nationality and recruited during their outpatient clinic visits. Cases with asthma were classified into subgroups according to disease severity, mild and severe asthma. Asthma patients were categorized according to Global Initiative for Asthma (GINA) management recommendation. Mild asthma patients fulfill step 1 and 2 GINA management recommendations. However, patients who fulfilled step 4 or 5 GINA management recommendations were categorized in severe asthma group.
Patients with mild asthma were generally characterized by infrequent symptoms and relatively well-preserved lung function, which minimally impact daily activities. In contrast, severe asthma involves persistent symptoms, frequent exacerbations, frequent use of corticosteroids course and significant limitations in lung function that often require intensive treatment and medical intervention.18,19
Ethics Approval and Consent to Participate
Ethical approval has been obtained from Kuwait University and the Ministry of Health, in accordance with the Helsinki Declaration protocol (Research study number MI02/22, 2022/2010) to ensure that the research is conducted ethically and in compliance with internationally recognized standards. Informed consent has been obtained from all participants involved in the study, as well as their legal guardians, to ensure that they are fully aware of the nature and purpose of the research and have given their voluntary and informed consent to participate.
Sample Size
The sample size was determined using Minitab 17.1.0.0 for Windows software (Minitab Inc., 2013, Pennsylvania, USA). A type I error of 0.05 and a type II error of 0.2 were considered. Based on Hijazi et al,20 the proportion of the IL4 gene -C590T polymorphism in the non-disease cohort was 0.76, and the odds ratio (OR) was 4. Therefore, the minimum calculated sample size would be 170 for a control: case ratio of 1:1 (85 in each limb) and 183 for a control: case ratio of 2:1 (minimum case=61 and minimum control=122). Moreover, considering Hong et al,21 assuming an odds ratio of 2, a disease prevalence of about 1%, a minor allele frequency of 5%, complete linkage disequilibrium (LD), and a 5% error rate in an allelic test to achieve 80% power, the minimum total sample size required would be 248 with a 1:1 case/control ratio. Additionally, to achieve a more robust representation of the subgroup of cases with varying degrees of asthma severity (mild and severe), an equal number of mild asthma cases would be needed to achieve 80% study power, assuming a case: case ratio similar to the control: case ratio.
Sample Collection
About 10 mL of venous blood was collected from all participants under a complete aseptic condition in a plastic syringe and then divided into two tubes; the first tube was without anticoagulant, and the other was with EDETA for DNA extraction. Samples were centrifuged at 4000 g for 10 min to separate the serum plasma and buffy coat containing leukocytes and then frozen at –20 ° C for DNA extraction. Genomic DNA extraction was performed from 200 μLblood using the Biospin Whole Blood Genomic DNA Extraction Kit (GentraPuregeneQIAGEN, USA). DNA concentration and purity were analyzed using the NanoDrop™ 1000 Spectrophotometer.
Genotyping
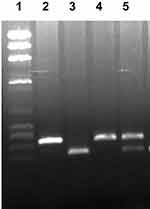
The primers and procedures used to determine IL4 gene -C590T, rs2243250 promoter polymorphism, has been described earlier.20 In which, the genotypes were determined in at least two independent readings to enhance accuracy, and as an additional quality control measure, DNA sequencing of the PCR products was carried out in selected samples. These measures were implemented to ensure the correctness and reliability of the identified genotypes. The CàT transition at codon 590 of the IL4 gene promoter region abolished a restriction site for BsmF1 in the T-allele. The polymorphism was detected by BsmF1 restriction endonuclease digestion of the PCR-amplified product. Agarose gel electrophoresis was used to analyze the cleavage products. Following agarose gel electrophoresis, the cleavage products were detected after staining with Ethidium bromide under UV light (Figure 1). The expected product sizes were 192 and 60 bp for the CC genotype, 252 bp for the TT genotype, and products of 252, 192, and 60 bp for the heterozygous genotype (CT) genotype.
Statistical Analysis
Demographic data of patients and control groups were collected in an Excel sheet, along with the genotype of each of the study participant. Minitab for Windows (Minitab Inc, 2013, version 17.1.0.0, Pennsylvania, USA) was used for statistical analysis. The data is represented as mean and standard deviation for numerical data and number (%) for categorical data.
For the dominant model, we calculated it by considering individuals who carried at least one copy of the variant allele as “affected” or “cases”, while those without any copies of the variant allele were designated as “unaffected” or “controls”. This approach allowed us to examine the impact of the variant allele in a binary manner. For the recessive model, we designated individuals as “affected” or “cases” only if they carried two copies of the variant allele. Those with one or no copies of the variant allele were categorized as “unaffected” or “controls” in this model.Comparison between two means was performed using an independent t-test and between two or more frequencies through a Chi-square test. Multiple comparisons were corrected using Bonferroni correction methods; the adjusted alpha level was 0.01 for significant threshold.
Logistic regression analysis with adjusted and non-adjusted methods was applied to find the predictive ability of IL-4 gene polymorphism and severe asthma. All tests were two-sided, and p<0.05 was considered significant for simple comparison and below 0.01 after correction for multiple comparison.
Results
IL-4 (C590T) Gene Polymorphism in Mild and Severe Subgroup of Asthma
Descriptive data of asthma cases and control were presented in Table 1; both were matched in sex but were significantly younger than the asthmatic patients, p=0.71 and <0.001, respectively.
 |
Table 1 Demographic Data of the Studied Groups |
Table 2 consisted of the three different genetic models used to investigate an association between IL-4 gene polymorphism and the risk of mild (102 cases) and severe asthma (113); the CC genotype was protective against mild asthma; its frequency was found to be significantly higher in the control group and severe asthma subgroup, p< 0.001 and 0.01, respectively. However, there was an insignificant difference between the control group and severe asthma, p=0.08. On the other hand, the CT genotype was significantly linked to mild asthmatic patients, p<0.001, and the likelihood of mild asthma increased threefold with this genotype. In contrast with the two polymorphisms mentioned above, the TT genotype did not show any significant association with any of the groups. Moreover, the frequency of the C allele was higher than the T alleles in control, mild, and severe asthmatic subgroups. In the control group and severe asthma subgroup, the frequency of the C allele was significantly higher than in the mild asthma subgroup: 78% versus 62% and 73% versus 62%, respectively, p=0.001 and 0.003. However, the control and severe asthma subgroups were matched with insignificant differences, p=0.04.
 |
Table 2 IL-4 (C590T) Gene Polymorphism in Mild and Severe Asthma |
In contrast, the frequency of the T allele was significantly higher in the mild asthmatic group compared with both the control group and severe asthma subgroup: 38% versus 22% and 38% versus 27%, respectively, p=0.002 and <0.001. Therefore, the T allele can be considered the risk allele for the associated risk of mild bronchial asthma.
No significant difference was detected between the three groups in the dominant model, p=0.23, 0.99, and 0.35, respectively. However, in the recessive model, the risk of mild asthma increased threefold compared to the control group and twofold to the severe asthma subgroup, p=0.001 and 0.01, respectively.
Risk Factors and Predictors of Severe Bronchial Asthma
The data on other risk factors and predictors of severe bronchial asthma are presented in Table 3. It showed that patients with severe bronchial asthma were significantly older than mild asthma patients, p<0.001. Additionally, they had an adult-onset disease and associated nasal polyp (NP), p=0.01 and < 0.001, respectively. In contrast, the frequency of oral corticosteroid (OCS) use was significantly lower, p=0.04, amongst the severe asthma patients.
 |
Table 3 Risk Factors Associated with Severe Asthma |
The predictors of severe asthma were summarized in Table 4, in which the recessive genetic model of the IL-4 gene decreased the risk of severe asthma in the non-adjusted model, OR=0.5, p=0.01. However, adjustment of IL-4 gene polymorphism with age, sex, and associated risk factors showed that the risk of severe bronchial asthma was unaffected by the IL-4 gene polymorphism, OR=0.92, p=0.80. The risk factors associated with developing severe asthma were being an elderly female with a history of childhood-onset disease and associate NP, OR=1.08, 2.01, 2.36, and 8.42; p<0.001, 0.05, 0.05, and <0.001, respectively. On the other hand, OCS use decreased the risk of severe asthma, OR=0.09, p=0.006.
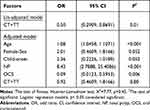 |
Table 4 Predictors of Severe Asthma |
Discussion
The current study was the first of her kind; it highlighted the difference between mild and severe asthma in correlation with IL-4 (C590T) gene polymorphism. The data showed that mild and severe asthma was different in associated risk factors and genetic risk. The frequency of C alleles in severe asthma was significantly higher than in mild asthma, and the risk of severe asthma increased twice with the CC genotype; it was protective against mild asthma but not severe asthma. Additionally, despite the overall frequency of C alleles being higher than T alleles in both asthmatic subgroups and control, the T alleles were the associated risk for mild asthma. A supporting study by Zhang et al22 stated that the IL-4-590C>T polymorphism might be linked to bronchial asthma in Uyghur children, and the T allele was more frequent in asthmatic Uyghur children; the study established that the genetic variation leads to increase IgE level and airway obstruction (FEV1%). Childhood asthma differs from adult asthma in its pathogenesis, as it is influenced by underdeveloped immune responses and the continued growth of lung function. Typically, childhood asthma is associated with allergic reactions to various allergens, such as pollens. This type of asthma is usually mild to moderate in severity and can be controlled effectively with proper medication adherence.23
An older Kuwaiti study20 denied any association between adult asthma and IL-4 (C590T) gene polymorphism, even with a different phenotypic classification of asthma (hay fever, eczema, and positive skin prick test group), in addition to finding similar frequencies of C alleles and T alleles in all studied groups (asthmatic, non-asthmatic and control). This discrepancy between the present data and the older study could be related to the difference in patient selection; Hijazi et al20 focused on patients with uncontrolled asthma and all cases collected from the hospital during an exacerbation; however, the present study was tented to be community-based style; the collection of cases and control happened during routine outpatient’s clinic visits. Another study found that asthma susceptibility is linked to the C allele and CC genotype of the IL-4-590C >T gene polymorphism and considered that a possible cause of asthma.23 Additionally, multiple studies suggested that a higher frequency of T alleles in control may be protective against developing asthma.24–26
To better understand the connection between the IL-4-590C>T gene polymorphism and asthma, a recent meta-analysis conducted a subgroup analysis. However, it was found that there was no significant association. This could be due to various factors, such as differences in sample size, ethnicity, source of control sample, and genotyping methods.27 While genetic and environmental factors both play a role in the development of asthma, there is still much to learn about genetic variation and severe asthma.28 Previous research has suggested that asthma is caused by multiple genes rather than just one gene having a significant impact.27
A meta-analysis found that specific variations (SNPs) in the IL4 gene are linked to asthma susceptibility in Caucasian-European populations, especially SNPs like rs2070874 (C-33T) and rs2243250 (C-589T), which are located in the gene’s promoter region. Notably, rs2070874 is one of the most frequently reported SNPs associated with asthma risk, especially in Caucasian and Asian populations, and it’s particularly tied to the severity of the condition.29
This study emphasized the importance of host-related risks in the development of severe asthma, in addition to the genetic variability of the IL-4-590C >T gene. The CT+TT genotype was found to be protective against severe asthma in a recessive genetic model, but it increased the risk of mild asthma by twice as much. However, other risk factors were found to play a more significant role in predicting severe asthma, rather than the IL-4-590C >T gene polymorphism. Elderly females were found to be twice as likely to develop severe asthma based on an adjusted regression analysis model. A Korean national database study showed that asthma was more severe in elderly females and associated with higher morbidity and mortality.30 The underlying cause is still unclear, but some studies suggest that sex hormones like estrogen and progesterone may contribute to the clinical features of asthma.31–33 Further investigation is needed to clarify the underlying mechanisms of severe asthma in elderly females, but the variation of IL-4-590C >T gene polymorphism could be considered a contributing factor.
The current data showed that the frequency of NP was higher in severe asthmatic patients than in mild asthma, and the coexistence of NP increased the risk of severe asthma eightfold more. In concordance with meta-analysis, NP was independently associated with severe asthma.34 Severe asthma and nasal polyps frequently coexist in some individuals, often called “asthma with nasal polyps”.35 Chronic inflammation is the characteristic hallmark of asthma and NP; it is characterized by overactive immune responses and an imbalance in specific cytokines, particularly IL-4, IL-5, and IL-13, which promote inflammation, tissue remodeling, and eosinophilic response.35,36
Mohammadi et al found a significant association between TT and CT genotypes at the IL-4 C590T promoter gene and NP.37 However, a contrary study from Korea showed that T alleles were protective against non-asthmatic and asthmatic nasal polyps.38 On the other hand, the Iranian study reported a lack of association between the IL-4 C590T promoter gene and NP.39
A longitudinal study that followed individuals with severe asthma for 10 years found that 83% of those who experienced severe asthma symptoms between the ages of 14 and 55 tended to respond well to their medication. However, the risk of persistent severe asthma was linked to other factors like low socioeconomic status, a high number of other health conditions, and non-adherence to medication during the first year of diagnosis.40 Surprisingly, sex and typical risk factors for childhood asthma did not play a role in the continued presence of severe asthma in this group.40
Our data showed that the frequency of adult-onset emergence asthma was significantly severe, even though, for those who started their asthma in childhood and persisted till adulthood, the risk of severe asthma increased twice.Asthma symptoms can last throughout childhood, adolescence, and adulthood for some people. However, many children with asthma show improvement and fewer symptoms during their school years.41 Boys with a milder and less allergic form of the disease tend to have a better prognosis.42 Nonetheless, it’s not clear if symptom remission means that the underlying problem has disappeared, as airway hyper-responsiveness and inflammation can persist even without visible symptoms.41 Some women with impaired lung function can develop asthma symptoms in adulthood,43 but it’s uncertain whether this represents a new onset or a relapse of childhood asthma. Both relapse and adult-onset asthma symptoms have been linked to allergic disease, sensitization, and airway hyper-responsiveness.44 Smoking has been associated with the persistence, relapse, and new onset of asthma symptoms in adulthood,45,46 although the underlying mechanisms are not fully understood.
Even though the study had an appropriate sample size, it had some limitations. The most important one is that it only discussed one genetic variation, while asthma pathogenesis involves multiple and complex genetic factors. Therefore, one gene-variability could not fully explain the association. Additionally, the researchers were unable to measure the level of IL-4 in serum for all groups studied, which could have reflected the gene’s variability, expression, and function in each group.
However, the study’s strength came from its stratification of adult asthma by severity and exploration of host-related risk factors along with genetic variability.
In conclusion, the data showed that T alleles, CT genotype in the co-dominant genetic model, and (CT+TT) genotype in the recessive model were significantly associated with mild asthma, not severe asthma. The IL-4 (C590T) gene polymorphism did not independently affect the risk of severe asthma. However, elderly females with a history of childhood-onset disease and associated nasal polyps had an increased risk of severe asthma up to 8 times.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Acknowledgement
The authors would like to acknowledge Dr. Ahmed Maher for his contributions to data collection.
Funding
This research received funding from Kuwait University Research Sector, project number (M102/22).
Disclosure
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Stern J, Pier J, Litonjua AA. Asthma epidemiology and risk factors. Semin Immunopathol. 2020;42(1):5–15. PMID: 32020334. doi:10.1007/s00281-020-00785-1
2. Ahmed H, Turner S. Severe asthma in children-a review of definitions, epidemiology, and treatment options in 2019. Pediatr Pulmonol. 2019;54(6):778–787. PMID: 30884194. doi:10.1002/ppul.24317
3. Devani P, Lo DKH, Gaillard EA. Practical approaches to the diagnosis of asthma in school-age children. Expert Rev Respir Med. 2022;16(9):973–981. PMID: 36125212. doi:10.1080/17476348.2022.2126355
4. Harun NS, Lachapelle P, Douglass J. Thunderstorm-triggered asthma: what we know so far. J Asthma Allergy. 2019;12:101–108. PMID: 31190900; PMCID: PMC6512777. doi:10.2147/JAA.S175155
5. Cecchi L, Vaghi A, Bini F, Martini M, Musarra A, Bilò MB. From triggers to asthma: a narrative review on epithelium dysfunction. Eur Ann Allergy Clin Immunol. 2022;54(6):247–257. PMID: 36214074. doi:10.23822/EurAnnACI.1764-1489.271
6. Mims JW. Asthma: definitions and pathophysiology. Int Forum Allergy Rhinol. 2015;5(1):S2–6. PMID: 26335832. doi:10.1002/alr.21609
7. Zinellu E, Piras B, Ruzittu GGM, Fois SS, Fois AG, Pirina P. Recent advances in inflammation and treatment of small airways in asthma. Int J Mol Sci. 2019;20(11):2617. PMID: 31141956; PMCID: PMC6601314. doi:10.3390/ijms20112617
8. Steinke JW, Borish L. Th2 cytokines and asthma. Interleukin-4: its role in the pathogenesis of asthma, and targeting it for asthma treatment with interleukin-4 receptor antagonists. Respir Res. 2001;2(2):66–70. PMID: 11686867; PMCID: PMC59570. doi:10.1186/rr40
9. Pawankar R, Okuda M, Yssel H, Okumura K, Ra C. Nasal mast cells in perennial allergic rhinitics exhibit increased expression of the Fc epsilonRI, CD40L, IL-4, and IL-13, and can induce IgE synthesis in B cells. J Clin Invest. 1997;99(7):1492–1499. PMID: 9119992; PMCID: PMC507968. doi:10.1172/JCI119311
10. Dabbagh K, Takeyama K, Lee HM, Ueki IF, Lausier JA, Nadel JA. IL-4 induces mucin gene expression and goblet cell metaplasia in vitro and in vivo. J Immunol. 1999;162(10):6233–6237. PMID: 10229869.
11. Shirakawa I, Deichmann KA, Izuhara I, Mao I, Adra CN, Hopkin JM. Atopy and asthma: genetic variants of IL-4 and IL-13 signalling. Immunol Today. 2000;21(2):60–64. PMID: 10652462. doi:10.1016/s0167-5699(99)01492-9
12. Ayakannu R, Abdullah NA, Radhakrishnan AK, Lechimi Raj V, Liam CK. Relationship between various cytokines implicated in asthma. Hum Immunol. 2019;80(9):755–763. PMID: 31054782. doi:10.1016/j.humimm.2019.04.018
13. Kumar P, Sharma DK, Ashawat MS. Pathophysiology and management of atopic dermatitis: a laconic review. Curr Drug Ther. 2020;15(4):321–336. doi:10.2174/1574885514666190828152316
14. Hook S, Cheng P, Holloway J, et al. Analysis of two IL-4 promoter polymorphisms in a cohort of atopic and asthmatic subjects. Exp Clin Immunogenet. 1999;16(1):33–35. PMID: 10087404. doi:10.1159/000019094
15. Rosenwasser LJ, Borish L. Genetics of atopy and asthma: the rationale behind promoter-based candidate gene studies (IL-4 and IL-10). Am J Respir Crit Care Med. 1997;156(4 Pt 2):S152–5. PMID: 9351597. doi:10.1164/ajrccm.156.4.12tac-14
16. Noguchi E, Shibasaki M, Arinami T, et al. Evidence for linkage between asthma/atopy in childhood and chromosome 5q31-q33 in a Japanese population. Am J Respir Crit Care Med. 1997;156(5):1390–1393. PMID: 9372650. doi:10.1164/ajrccm.156.5.9702084
17. Jin X, Zheng J. IL-4-C-590T locus polymorphism and susceptibility to asthma in children: a meta-analysis. J Pediatr. 2021;97(3):264–272. PMID: 32781035; PMCID: PMC9432276. doi:10.1016/j.jped.2020.05.005
18. Louis R, Satia I, Ojanguren I, et al. European respiratory society guidelines for the diagnosis of asthma in adults. Eur Respir J. 2022;15:2101585. PMID: 35169025. doi:10.1183/13993003.01585-2021
19. Ibrahim MA, Ismail AI, Rani MF. A brief review of severe asthma. J Res Health Sci. 2021;6(2):4–12. doi:10.24191/jchs.v6i2.14942
20. Hijazi Z, Haider MZ. Interleukin-4 gene promoter polymorphism [C590T] and asthma in Kuwaiti Arabs. Int Arch Allergy Immunol. 2000;122(3):190–194. doi:10.1159/000024396
21. Hong EP, Park JW. Sample size and statistical power calculation in genetic association studies. Genomics Inform. 2012;10(2):117–122. PMID: 23105939; PMCID: PMC3480678. doi:10.5808/GI.2012.10.2.117
22. Zhang JH, Zhang M, Wang YN, Zhang XY. Correlation between IL-4 and IL-13 gene polymorphisms and asthma in Uygur children in Xinjiang. Exp Ther Med. 2019;17(2):1374–1382. PMID: 30680016; PMCID: PMC6327510. doi:10.3892/etm.2018.7096
23. Pijnenburg MW, Frey U, De Jongste JC, Saglani S. Childhood asthma: pathogenesis and phenotypes. Eur Respir J. 2022;59(6):2100731. PMID: 34711541. doi:10.1183/13993003.00731-2021
24. Hussein IA, Jaber SH. Genotyping of IL-4 −590 (C>T) Gene in Iraqi Asthma Patients. Dis Markers. 2017;2017:5806236. PMID: 28386156; PMCID: PMC5366214. doi:10.1155/2017/5806236
25. Berenguer AG, Fernandes AT, Oliveira S, et al. Genetic polymorphisms and asthma: findings from a case-control study in the Madeira island population. Biol Res. 2014;47(1):40. PMID: 25299150; PMCID: PMC4167518. doi:10.1186/0717-6287-47-40
26. Smolnikova MV, Smirnova SV, Freidin MB, Tyutina OS. Immunological parameters and gene polymorphisms (C-590T IL4, C-597A IL10) in severe bronchial asthma in children from the Krasnoyarsk region, West Siberia. Int J Circumpolar Health. 2013;72:21159. PMID: 23984295; PMCID: PMC3753143. doi:10.3402/ijch.v72i0.21159
27. Liu T, Yin W, Luo L, Wu Y, Qin S, Qin X. Association between Interleukin-4-590C>T polymorphism and the susceptibility to asthma: a meta-analysis of case-control study. J Healthc Eng. 2022;2022:1712715. PMID: 35392151; PMCID: PMC8983229. doi:10.1155/2022/1712715
28. Morales E, Duffy D. Genetics and gene-environment interactions in childhood and adult onset asthma. Front Pediatr. 2019;7:499. PMID: 31921716; PMCID: PMC6918916. doi:10.3389/fped.2019.00499
29. Tang L, Lin HG, Chen BF. Association of IL-4 promoter polymorphisms with asthma: a meta-analysis. Genet Mol Res. 2014;13(1):1383–1394. PMID: 24634237. doi:10.4238/2014.February.28.11
30. Park SY, Kim JH, Kim HJ, et al. High prevalence of asthma in elderly women: findings from a Korean national health database and adult asthma cohort. Allergy Asthma Immunol Res. 2018;10(4):387–396. PMID: 29949835; PMCID: PMC6021593. doi:10.4168/aair.2018.10.4.387
31. Baptist AP, Hamad A, Patel MR. Special challenges in treatment and self-management of older women with asthma. Ann Allergy Asthma Immunol. 2014;113(2):125–130. PMID: 25065349; PMCID: PMC4153393. doi:10.1016/j.anai.2014.05.013
32. Balzano G, Fuschillo S, De Angelis E, Gaudiosi C, Mancini A, Caputi M. Persistent airway inflammation and high exacerbation rate in asthma that starts at menopause. Monaldi Arch Chest Dis. 2007;67(3):135–141. PMID: 18018752. doi:10.4081/monaldi.2007.484
33. Zemp E, Schikowski T, Dratva J, Schindler C, Probst-Hensch N. Asthma and the menopause: a systematic review and meta-analysis. Maturitas. 2012;73(3):212–217. PMID: 22964072. doi:10.1016/j.maturitas.2012.08.010
34. Laidlaw TM, Mullol J, Woessner KM, Amin N, Mannent LP. Chronic rhinosinusitis with Nasal Polyps and asthma. J Allergy Clin Immunol Pract. 2021;9(3):1133–1141. PMID: 33065369. doi:10.1016/j.jaip.2020.09.063
35. Birs I, Boulay ME, Bertrand M, Côté A, Boulet LP. Heterogeneity of asthma with nasal polyposis phenotypes: a cluster analysis. Clin Exp Allergy. 2023;53(1):52–64. PMID: 36317421. doi:10.1111/cea.14247
36. Akdis CA, Bachert C, Cingi C, et al. Endotypes and phenotypes of chronic rhinosinusitis: a PRACTALL document of the European Academy of allergy and clinical immunology and the American Academy of allergy, asthma & immunology. J Allergy Clin Immunol. 2013;131(6):1479–1490. PMID: 23587334; PMCID: PMC4161279. doi:10.1016/j.jaci.2013.02.036
37. Mohammadi M, Dabiri S, Mollaei HR, et al. C-590T promoter polymorphism of the Interleukin (IL)-4 gene is associated with an increased usceptibility to nasal polyposis. Rep Biochem Mol Biol. 2019;7(2):129–135. PMID: 30805391; PMCID: PMC6374060.
38. Yea SS, Yang YI, Park SK, et al. Interleukin-4 C-590T polymorphism is associated with protection against nasal polyps in a Korean population. Am J Rhinol. 2006;20(5):550–553. PMID: 17063753. doi:10.2500/ajr.2006.20.2936
39. Nikakhlagh S, Ghadiri A, Saki N, Kardoni M, Konari A. Evaluation of C-590T Promoter of IL-4 gene polymorphisms in patients with sinonasal polyposis. Int J Pharm Res Allied Sci. 2016;5(2):39–43.
40. Chen W, Marra CA, Lynd LD, FitzGerald JM, Zafari Z, Sadatsafavi M. The natural history of severe asthma and influences of early risk factors: a population-based cohort study. Thorax. 2016;71(3):267–275. PMID: 26732738. doi:10.1136/thoraxjnl-2015-207530
41. Trivedi M, Denton E. Asthma in children and adults-what are the differences and what can they tell us about asthma? Front Pediatr. 2019;7:256. PMID: 31294006; PMCID: PMC6603154. doi:10.3389/fped.2019.00256
42. Schatz M, Rosenwasser L. The allergic asthma phenotype. J Allergy Clin Immunol Pract. 2014;2(6):645–8; quiz 649. PMID: 25439351. doi:10.1016/j.jaip.2014.09.004
43. Naeem A, Silveyra P. Sex differences in paediatric and adult asthma. Eur Med J. 2019;4(2):27–35. PMID: 31328173; PMCID: PMC6641536. doi:10.33590/emj/10312930
44. Christou EAA, Giardino G, Stefanaki E, Ladomenou F. Asthma: an undermined state of immunodeficiency. Int Rev Immunol. 2019;38(2):70–78. PMID: 30939053. doi:10.1080/08830185.2019.1588267
45. Dharmage SC, Perret JL, Custovic A. Epidemiology of asthma in children and adults. Front Pediatr. 2019;7:246. PMID: 31275909; PMCID: PMC6591438. doi:10.3389/fped.2019.00246
46. Fuchs O, Bahmer T, Rabe KF, von Mutius E. Asthma transition from childhood into adulthood. Lancet Respir Med. 2017;5(3):224–234. PMID: 27666650. doi:10.1016/S2213-2600(16)30187-4
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.