Back to Journals » Infection and Drug Resistance » Volume 17
Inhibiting Peptidoglycan Hydrolase Alleviates MRSA Pneumonia Through Autolysin-Mediated MDP-NOD2 Pathway
Authors Yang Y, Yao Z, Zhang J, Shao W, Li B , Wu H, Tang W, Zhang J
Received 17 December 2023
Accepted for publication 15 March 2024
Published 26 March 2024 Volume 2024:17 Pages 1231—1242
DOI https://doi.org/10.2147/IDR.S455339
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Sandip Patil
Yang Yang,1,2 Zongze Yao,1 Jiazhen Zhang,1 Wei Shao,3 Bo Li,2 Huihui Wu,2 Wenjian Tang,3 Jing Zhang2
1School of Medicine, Anhui University of Science and Technology, Huainan, People’s Republic of China; 2Anhui Province Key Laboratory of Occupational Health, Anhui No.2 Provincial People’s Hospital, Hefei, People’s Republic of China; 3School of Pharmacy, Anhui Medical University, Hefei, People’s Republic of China
Correspondence: Wenjian Tang; Jing Zhang, Tel +86-551-63672601, Email [email protected]; [email protected]
Background: Methicillin-resistant Staphylococcus aureus (MRSA) is a cause of staph infection that is difficult to treat because of resistance to some antibiotics. A recent study indicated that diarylurea ZJ-2 is a novel antibacterial agent against multi-drug resistant Enterococcus faecium. In this work, we refined the bactericidal mechanism of ZJ-2 as a peptidoglycan (PG) hydrolase by affecting AtlA-mediated PG homeostasis.
Methods: A wild-type strain (WT) and a mutant strain (ΔatlA) were used to investigate the effects of ZJ-2 on the cell wall, PG, and autolysin regulatory system by antimicrobial susceptibility testing, hemolytic toxin assay, microanalysis, autolysis assay, qRT-PCR, ELISA and mouse model of pneumonia.
Results: The results revealed that ZJ-2 down-regulated the expression of genes related to peptidoglycan hydrolase (PGH) (sprX, walR, atlA, and lytM), and reduced the levels of PG, muramyl dipeptide (MDP), cytokines, and hemolytic toxin, while ΔatlA interfered with the genes regulation and PG homeostasis. In the mouse MRSA pneumonia model, the same trend was observed in the nucleotide oligomerization domain protein 2 (NOD2) and relative proinflammatory factors.
Conclusion: ZJ-2 may act as a novel inhibitor of PG hydrolyse, disrupting autolysin-mediated PG homeostasis, and reducing inflammation by down-regulating the MDP-NOD2 pathway.
Keywords: MRSA, peptidoglycan hydrolase, homeostasis, autolysis, NOD2
Graphical Abstract:
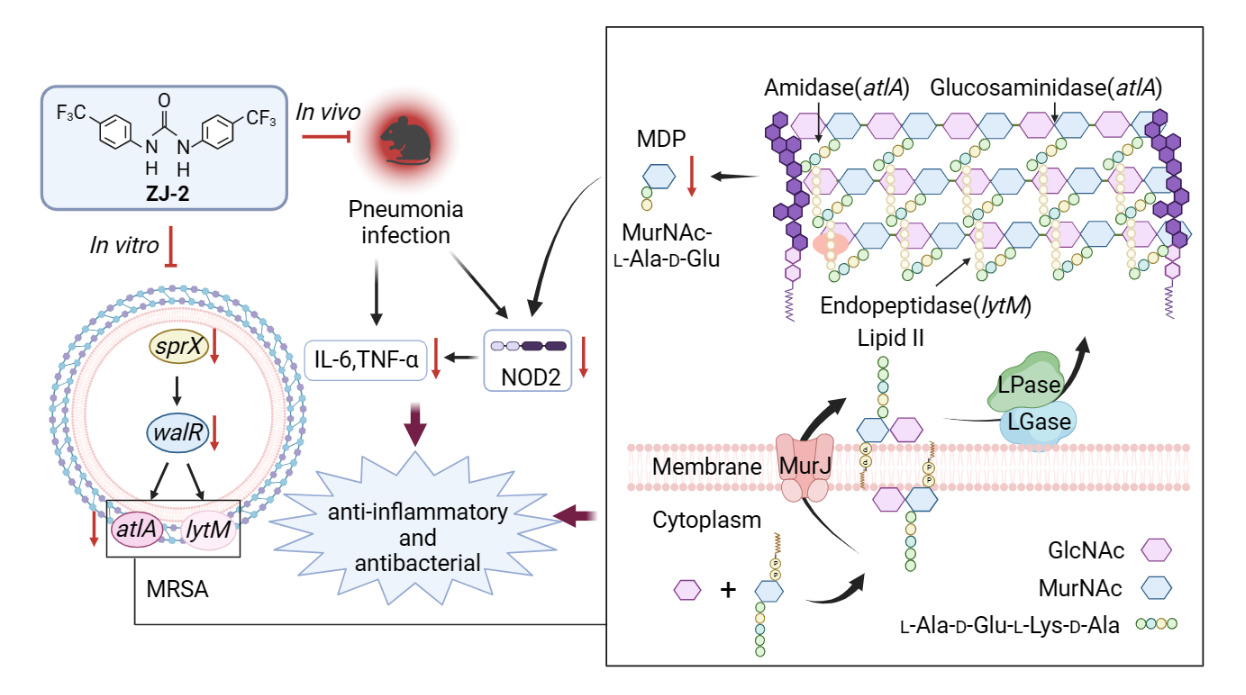
Introduction
Staphylococcus aureus (S. aureus) is a Gram-positive pathogen that causes a wide range of infections and diseases including sepsis, pneumonia, and skin infections.1,2 In the wake of the COVID-19 global pandemic, the misuse of antibiotics has exacerbated the situation of microbial resistance.3 MRSA is one of the common and genotypically diverse drug-resistant bacteria with very different heritabilities between carriers and infected individuals.4,5 The cell walls of gram-positive bacteria are composed of tough and resilient PG that maintain cell shape, resist swelling pressures inside the bacteria, and protect the cell from external mechanical damage.6,7 PG consists of relatively short glycan chains which are highly cross-linked via pentaglycine bridges, forming a highly dynamic structure.8,9 Bacteria not only synthesize and modify new PG networks but also maintain homeostasis between old and new PGs through cleavage and turnover.10 Antibiotics like penicillin and vancomycin inhibit PG synthesis, making it a preferred target for antibiotic development.11,12 The regulation of cell wall hydrolysis can lead to cell lysis and separation.13
Bacterial autolysin is responsible for regulating the expression of PGH in the cell wall to maintain cell homeostasis. When there are changes in PGH activity, the balance between PG synthesis and hydrolysis is disrupted, resulting in cell autolysis and death.14 The autolysin regulatory system involves various proteases and genes. In S. aureus, PGHs are regulated by an essential two-component system called WalRK.15 The upstream gene sprX not only positively regulates the expression of different autolysins through the regulator walR but also influences the secretion of MRSA proteins and virulence factors such as δ-hemolysin.16,17
AtlA, the major PGH involved in cell wall homeostasis, plays a crucial role in the development of biological membranes and cell wall metabolism.18 AtlA also acts as a virulence factor in bacterial biofilm formation by facilitating cell adhesion. Inhibition of AtlA secretion prevents biofilm production and eliminates existing biofilms. Therefore, the regulation of AtlA during cell wall lysis is more complex in Gram-positive bacteria.19,20
A recent study demonstrated that diarylurea ZJ-2 (Figure 1) can break down bacterial cell walls and decrease initial biofilm adhesion. This is achieved by suppressing the expression of PGHs genes (sagA and atlA) in Enterococcus faecium E3101.21 To understand the molecular mechanism of ZJ-2 on autolysin-mediated PGHs, we generated the atlA mutant strain to compare the wild strain MRSA USA300. Similar to cells treated with ZJ-2, the loss of atlA function resulted in reduced bacterial growth, autolysis activity, toxin secretion, and pharmacodynamics. Furthermore, using a mouse model of MRSA pneumonia, we discovered that ZJ-2 displayed a bactericidal effect by targeting AtlA and influencing PG homeostasis and subsequent immunoregulation.
 |
Figure 1 Schematic chemical structure formula of diarylurea ZJ-2. |
Materials and Methods
Chemicals and Bacterial Strains
MRSA strain USA300 (WT) and its mutant strain after it knocked out gene atlA (ΔatlA) were grown in Tryptic Soy Broth (TSB, Coolaber Science&Technology Co., Ltd. China) medium at 37°C. The reagents included ZJ-2 (1,3-bis(4-(trifluoromethyl)phenyl)urea), vancomycin (Solarbio Science&Technology Co., Ltd. China), Miller-Hinton broth (MHB, Thermo Fisher Scientific Inc. USA), Tris-Cl, Triton X-100 (Labgic Science&Technology Co., Ltd. China), Bacteria PG ELISA kit (Chuzhou shinuoda Biological Technology Co., Ltd. China), MDP ELISA kit (Nanjing SenBeiJia Biological Technology Co., Ltd. China), Mouse IL-6 and TNF-α ELISA kit (Elabscience Biotechnology Co., Ltd. China).
Construction of atlA Knockdown Strain
The deletion mutant was constructed by homologous recombination using plasmid pBT2 as described earlier.22,23 A spectinomycin selection marker (spc) which is flanked by upstream and downstream regions of the gene atlA in the cloning site of pBT2 was cloned. PCR amplification was performed using primers described in Table 1 and cloned in the antisense orientation into pBT2 plasmid at BamHI/EcoRI site. This knockout plasmid was introduced into S. aureus RN4220 by electroporation, and subsequently into MRSA USA300. The deletion mutant (ΔatlA: spc) obtained after homologous recombination was identified by its chloramphenicol-sensitive and spectinomycin-resistant phenotype and was verified by DNA sequencing.
 |
Table 1 Cloning and qRT-PCR Primers in This Study |
Minimum Inhibitory Concentration (MIC) Assay
MIC assay was performed using the MHB dilution method as described by the Clinical and Laboratory Standards Institute (CLSI). Briefly, 100 μL of MHB was filled into each well of the 96-well microplate (Wuhan Servicebio Technology Co., Ltd. China). Certain concentrations of ZJ-2 and VAN were added to the first column of microwells, and diluted sequentially. The initial concentrations of ZJ-2 and VAN were 10 and 100 μmol/L, respectively. The first row of wells that were only added with MHB medium served as a blank control group. The last row of wells that were only added with MHB medium and MRSA USA300 served as the positive control group. The bacterial suspension of MRSA USA300 was diluted to 1‒5 × 105 cfu/mL. 96 microplate was incubated at 37°C for 24 h. MIC values were read using a spectrophotometer assay (Bio-Tek Epoch-2, USA). Three independent inoculation tests were used for all assays.
Growth Inhibition Assay
As mentioned previously, the assay was conducted to investigate the antimicrobial activity of MRSA USA300 and determine the inhibition of its growth with changes in dose and time. MRSA USA300 was inoculated into MHB medium containing different concentrations of agents, ranging from 5×105 cfu/mL. The concentrations of the agents used were 1/4MIC, 1/2MIC, MIC, and 2MIC, respectively. The plates were incubated at 37°C for 24 hours, and the OD values at 600 nm were measured at 4-hour intervals. Each sample was tested in triplicate.
Hemolysis Assay
Hemolysis was assayed as previously described with some modification.26,27 A hemolysis assay of ZJ-2 was carried out. Firstly, the rabbit blood was collected from the saphenous vein. Afterwards, the anticoagulated erythrocyte precipitate was obtained by centrifugation and washed 2–3 times with equal amounts of saline. After the supernatant was clear and transparent, it was prepared with saline to 10% rabbit erythrocytes. MRSA USA300 was treated with 0, MIC, 2MIC, 3MIC and 4MIC of ZJ-2. Samples were cultured overnight at 37°C, centrifuged at 12,000g for 2 min, then 100 μL of the supernatant was mixed with 875 μL PBS and 25 μL rabbit erythrocytes and incubated at 37°C for 30 min. Triton X-100 as a positive control, PBS as a negative control, and the mixture was centrifuged at 10000g and 4°C for 1 min. The hemolysis inhibition rate was calculated by measuring the absorbance value at OD543 with a microplate reader.
Electron Microscope
Electron microscope was assayed as previously described with some modifications.28 MRSA USA300 suspension was adjusted to 1.5×108 cfu/mL, added to TSB medium, treated with 0, 1/4MIC, 1/2MIC of ZJ-2, and incubated at 37°C for 24 h. Bacterial precipitate was collected by centrifugation at 4,000 rpm for 10 min and rinsed with PBS. Cells were then fixed with 2.5% (v/v) glutaraldehyde at 4°C for 16 h and further dehydrated through a series of graded alcohols (30, 50, 70, 90 and 100%) for 15 min. After serial dehydration, CM-10 TEM analysis was used to examine fixed cells at 80 kV operating voltage. The SEM model used is the S4800 at 3.0 kV.
Triton X-100-Induced Autolysis Assay
Triton X-100-induced autolysis assay was performed as specified previously with slight modification.19 MRSA USA300 was cultured overnight and inoculated in TSB medium at 1: 50. ZJ-2 (or VAN) was added and final concentrations were set to 0, 1/4MIC and 1/2MIC. The culture was incubated at 37°C for 3 h. The precipitate was collected by centrifugation and washed twice with pre-cooled saline. Then it was re-suspended in autolysis buffer (50 mM Tris-Cl, pH 7.2, 0.2% Triton X-100) and incubated at 37°C. 200 μL was taken every 1h to determine its OD600 value, and replicated 3 times. The autolysis rate was calculated at 0, 1, 2, 3, and 4 h.
RNA Extraction and qRT-PCR
qRT-PCR detected the expression levels of MRSA genes (Bio-Rad, Hercules, USA).29 The primers used for qRT-PCR are listed in Table 1.24,25 Total RNA was extracted from MRSA USA300 treated with ZJ-2 (late exponential phase, 24 h) using the TRIzol method for RNA extraction and converted to cDNA and using SuperScript™ III First-Strand Synthesis SuperMix for qRT-PCR (Invitrogen, USA), and quantified with a Light Cycler 96 (Roche, Switzerland). The quantitative PCR cycle threshold (CT) results were analyzed using the comparative CT method (2-ΔΔCT method) with some modifications. All samples were analyzed in triplicate.
ELISA Kits for PG and MDP
MRSA USA300 was cultured overnight and inoculated in TSB medium at 1: 50. The concentrations were set to 0, 1/4MIC and 1/2MIC after adding ZJ-2 and incubated to the logarithmic phase of growth at 37°C. The supernatant was collected by centrifugation at 4000g for 10 min as a test sample. Using the ELISA kits, the assay was performed according to the steps in the instruction manual, and all samples were analyzed in three replicates.
Mouse Pneumonia Model
Animals: SPF female C57BL/6J mice (6‒8 weeks old; 18‒20 g) were purchased from Hefei Qingyuan Biotechnology Co., Ltd; mice were maintained with SPF food and water for 1 week. The animal room was 20‒26°C, 40‒70% temperature and humidity, 12 h alternating day and night, and animals were fed normally before the experiment. Mice were randomly divided into 7 groups: WT ZJ-2 (5 mg/kg), WT VAN (5 mg/kg), WT bacterial control, ΔatlA ZJ-2 (5 mg/kg), ΔatlA VAN (5 mg/kg), ΔatlA bacterial control, and blank group (n = 8). 80 µL of MRSA USA300 suspension (3 × 108 cfu/mL) was added by tracheal drip. All four treatment groups of mice were injected intraperitoneally after 24 h. The bacterial control and blank groups were given the same dose of saline. Mice were administered every 24 h and euthanized after 7 d. Orbital venous blood was collected for white blood cells (WBC), neutrophils (NEUT) and lymphocytes (LY) counts. The alveolar lavage fluid was collected to determine the levels of IL-6 and TNF-α by ELISA. Moreover, lung tissues were collected, and subjected to HE staining, detected the expression levels of inflammatory cytokines and NOD2 by qRT-PCR (Table 1). VAN was used as a control in the mouse MRSA pneumonia model (Figure 2).
 |
Figure 2 Flowchart for MRSA-induced pneumonia infection model in mice. |
Statistical Analysis
All statistical analyses were performed using SPSS software (SPSS 13.0 for Windows). The data are presented as the mean ± SD (n = 3). Differences between means were evaluated by Student’s t-test and defined as significant at P < 0.05. *P < 0.05; **P < 0.01; ***P < 0.001.
Results
ZJ-2 Inhibited the Growth of MRSA USA300
Firstly, the assay showed that MIC values of ZJ-2 and VAN were 0.11 and 1.13 mg/L (0.31 and 0.78 μmol/L), respectively, thus ZJ-2 showed a stronger inhibition against MRSA than VAN. Secondly, Figure 3 displayed the time-kill curves of ZJ-2 and VAN against WT and ΔatlA. Both ZJ-2 (Figure 3A and B) and VAN (Figure 3C and D) at MIC had significant growth inhibitory effects on WT and ΔatlA. Finally, at 1/2MIC and 1/4MIC, ZJ-2 exhibited stronger inhibition against WT than against ΔatlA, but that was not the case for VAN. Therefore, the deletion of atlA can affect the inhibitory activity of ZJ-2 against MRSA.
ZJ-2 Inhibited the Secretion of MRSA USA300-Hemolytic Toxin
Hemolysis assay showed that ZJ-2 could significantly inhibit the hemolytic activity of MRSA in a dose-dependent manner (Figure 4A and B). Inhibition rates of hemolysis of WT and ΔatlA were 99.4%, 98.7%; 99.6%, 66.3%; 99.7%, 2.35% and 8.69%, 1.02% at 4MIC, 3MIC, 2MIC and MIC, respectively. Obviously, the deletion of atlA affected the anti-hemolysis of ZJ-2 against MRSA, therefore, ZJ-2 can regulate hemolytic toxin secretion through the expression of gene atlA.
Ultrastructural Analysis
Differences in the cell surface of WT and ΔatlA cells were observed by SEM. Compared with WT (Figure 5A and B), ΔatlA (Figure 5C and D) exhibited a rough cell surface with filamentous intercellular adhesions and thicker cell walls than WT. The effects of ZJ-2 on cell walls of WT and ΔatlA were different. After treatment with ZJ-2 at 1/4MIC, WT cells showed irregular septation, inhibition of division and thickened cell walls (Figure 5E), in contrast, ΔatlA appeared less altered, and the cell wall appeared slightly thickened (Figure 5F). After treatment with ZJ-2 at 1/2MIC, WT cell walls had high swelling pressures due to inhibition of division and constant expansion, the cell wall ruptured, the contents leaked, and the cells died (Figure 5G), inversely, ΔatlA cells were hardly affected, only observed more abnormal septa and less cell wall rupture (Figure 5H). In short, the effect of ZJ-2 on MRSA was dependent on the expression of atlA.
The Ability of ZJ-2 to Inhibit MRSA USA300 Autolysis
Triton X-100-induced autolysis experiments showed that ZJ-2 inhibited the autolysis of WT (from 63.0% to 48.0%) in a dose-dependent manner, while for ΔatlA, this change induced by ZJ-2 was very small (Figure 6A). Differently, VAN hardly affected the autolysis (from 62.1% to 65.5%) for WT and ΔatlA (Figure 6B). The results indicated that the autolysin regulation of ZJ-2 was different from that of VAN. VAN is known as a PG synthase inhibitor, but, ZJ-2 may target PGHs.
ZJ-2 Regulated the Expression of Autolysin-Related Genes
qRT-PCR was used to examine the expression of autolysin-related genes. SprX positively influences the WalR expression and in turn regulates the expression of AtlA and LytM. In WT, ZJ-2 treatment at 1/4 MIC exhibited down-regulation of multiple autolysins, and 0.28, 0.79, 0.79 and 0.75 reduction of sprX, walR, atlA, and lytM transcripts in qRT-PCRs, respectively (Figure 7A). As a contrast, in ΔatlA, sprX, walR and lytM transcripts were decreased by 0.69, 0.48 and 0.49, respectively (Figure 7B). After ZJ-2 treatment at 1/2MIC, more effective action and the same trend were observed. Knockout of the atlA influenced the differential expression of multiple autolysins, however, ZJ-2 regained antibacterial activity through the regulation of autolysis-mediated PG homeostasis. Therefore, the bactericidal effect of ZJ-2 acted by the autolysin regulation system controlling autolysis activity.
ZJ-2 Inhibited PG Cleavage and MDP Production in MRSA USA300
The inhibitory effect of ZJ-2 on PG hydrolysis was verified by detecting the content of free PG and MDP in the supernatant. The results showed that PG and MDP content in the supernatant of WT after 1/4MIC ZJ-2 incubation hardly changed, while, those after 1/2MIC ZJ-2 incubation decreased. Conversely, PG and MDP content in the supernatant of ΔatlA after ZJ-2 incubation did not change (Figure S1A and B). In all, ZJ-2 could inhibit PG hydrolysis and reduce MDP production by down-regulating the expression of autolysin-related genes.
Effect of ZJ-2 on Inflammatory Cells in the Blood
The therapeutic effect of ZJ-2 on MRSA infection was evaluated by detecting the number of inflammatory cells in the blood of mice. As shown in Figure S2, WBC, NEUT and LY were elevated in the blood of WT and ΔatlA infection mice. After treatment with ZJ-2 (or VAN), whether WT or ΔatlA, these cells were all reduced. Notably, the number of inflammatory cells in WT-infected mice exhibited a greater reduction than those in ΔatlA-infected mice. This further indicated that ZJ-2 reduced inflammatory responses of MRSA infection by inhibiting autolysin-mediated PG hydrolysis.
Effect of ZJ-2 on the Pathological Changes of Lung Tissue
The pathological changes in the lung tissue of mice were observed by H&E staining. As shown in Figure 8, in WT group, we observed extensive lung tissue damage, necrosis caused by inflammatory cell infiltration and alveolar collapse. After treatment with ZJ-2 (or VAN), the inflammatory cell infiltration of lung tissue was significantly reduced, and the alveolar collapse caused by infection gradually recovered. In ΔatlA group, the ability of ZJ-2 and VAN to restore lung tissue became weaker. The results suggested that the deletion of atlA reduces the therapeutic effect of ZJ-2.
 |
Figure 8 Lung histology stained with H&E. HE staining’s view is 50 μm. |
Effect of ZJ-2 on the Level of NOD2
qRT-PCR was used to detect changes in the transcript levels of the gene NOD2. For WT, both ZJ-2 group and VAN group reduced the transcript level of gene NOD2 (Figure 9A). However, for ΔatlA, the down-regulation was attenuated. This change may be due to the reduction of PG and MDP by atlA-mediated autolysin. Thus, ZJ-2 can alleviate inflammatory response by the down-regulation of MDP-NOD2 pathway including the autolysin atlA.
 |
Figure 9 qRT-PCR detected the expression of NOD2 (A), IL-6 (B) and TNF-α (C) in mouse lung tissues. Values represent means ± SD; ***P < 0.001, **P < 0.01, *P < 0.05, when compared with the control. |
Effect of ZJ-2 on the Levels of Cytokine
qRT-PCR (Figure 9B and C) and ELISA (Figure S3) were used to detect the levels of the inflammatory factors IL-6 and TNF-α in mouse alveolar lavage fluid and blood. Both ZJ-2 and VAN treatment decreased the levels of inflammatory factors, but the levels in ΔatlA showed a more significant decline than those in WT. This suggested that ZJ-2 can decrease the levels of relative inflammatory cytokines by inhibiting MDP-NOD2 pathway.
Discussion
In recent decades, there has been a significant focus on β-lactam antibiotics (such as cephalosporins) and VAN, which target the process of PG synthesis in the cell wall. However, after the global pandemic of COVID-19, due to the microbial resistance of the misuse of antibiotics, the sites of MRSA infection showed a high degree of diversification and some isolate types were more prevalent among carriers than at the site of infection.3–5 As a result, there is an urgent need to search for novel antibiotics and explore new antibacterial mechanisms. One promising target for drug development and immunization is PGHs.30 These enzymes, including glycosidases, amidases, and endopeptidases, are regulated by AtlA and LytM. AtlA secretes bifunctional multi-domain enzymes, namely N-acetylmuramyl-L-alanine amidase and endo-beta-N-acetylglucosaminidase.18,19 Multiple PGHs are interconnected to form a complex and tightly regulated autolysin system. When externally stimulated by antibiotics, this system is abnormally expressed and the expression of PGHs becomes dysregulated, making bacteria highly susceptible to cell wall lysis and autolysis.
PG homeostasis is a dynamic process in the cell wall that is essential for the growth (life) of S. aureus and the bactericidal effects of cell wall antibiotics (death).31 This process involves the synergistic action of PG synthesis and hydrolysis. When PG hydrolysis is normal but synthesis is slowed, an imbalance in PG biosynthesis occurs, leading to bacterial autolysis.13 The TritonX-100-induced autolysis assay revealed that treatment with ZJ-2 significantly inhibits autolysis, while VAN treatment did not show a noticeable change in autolytic capacity (Figure 6). SprX plays a positive role in the expression of WalR, which in turn regulates autolytic activity, particularly AtlA and LytM. Decreased expression of SprX and WalR reduces the virulence of MRSA.15,16 In this study, the expression of autolysin genes (sprX, walR, atlA, and lytM) was significantly suppressed. When gene atlA was deleted, ZJ-2 showed differential regulation of autolysins (Figure 7). Conversely, β-lactam antibiotics can promote bacterial autolysis by altering the proteolytic process,32,33 while ZJ-2 targets PGHs by interfering with PG homeostasis, resulting in dysregulation of the autolysin regulatory system and inducing bacterial cell wall expansion and death.
In addition, a thick cell wall aggravates antibiotic resistance. TEM observed the ultrastructural difference between WT and ΔatlA cell walls (Figure 5). When bacterial autolysin activity is lacking, PG cleavage is reduced,18 resulting in a rough and thickened cell wall surface. ZJ-2 treatment destroyed the cell wall ultrastructure in WT, but this change was less pronounced in ΔatlA. The mutation in ΔatlA decreased autolysis sensitivity by inhibiting transcription and synthesis, leading to increased resistance to β-lactam antibiotics against S. aureus.34,35 However, the spectrum of antibiotic resistance can vary among different staphylococcal lineages. The sagB mutant of MRSA showed increased sensitivity to the β-lactam antibiotic oxacillin due to an increase in glycan chain length, but it remained resistant to methicillin.36 Inactivation of AtlA weakened the inhibitory effect of ZJ-2 against MRSA, affecting MRSA growth, hemolytic toxins, and cell wall cleavage. Thus, knockdown of atlA increased resistance to ZJ-2 against MRSA (Figures 3 and 4).
MDP, a proinflammatory factor, stimulates the release of proinflammatory cytokines, playing a key role in host recognition and immune response stimulation.37 In the mammalian immune system, the nucleotide-binding NOD2 receptor recognizes PG and is mainly expressed in monocytes and epithelial cells.38 As autolytic activity decreases, the release of MDP by PG hydrolysis also decreases, leading to a subsequent reduction in the host inflammatory response.39 This study found that the deletion of atlA influenced PG hydrolysis, and ZJ-2 inhibited PG hydrolysis, resulting in a decrease in MDP levels through autolysin regulation (Figure 7 and Supplementary 1). In an in vivo study, it was confirmed that MDP aggravated damage in MRSA pneumonia, while ZJ-2 treatment reduced white blood cell infiltration, thereby inhibiting the MDP-NOD2 pathway and attenuating experimental pneumonia (Figures 8 and 9, Supplementary 2 and Supplementary 3).
Conclusion
In general, ZJ-2 plays a crucial role in both virulence and the host inflammatory response by regulating autolytic activity. Deletion of atlA resulted in increased resistance to MRSA in ZJ-2. Furthermore, ZJ-2 may serve as a promising inhibitor of PG hydrolase, disrupting autolysin-mediated PG homeostasis, thereby preventing cell division and PG hydrolysis. Additionally, ZJ-2 down-regulates the MDP-NOD2 pathway, leading to a reduction in inflammation. This study also presents a novel approach for the identification of antibacterial agents that target PG hydrolase.
Data Sharing Statement
The original contributions presented in the study are included in the article; further inquiries can be directed to the corresponding authors.
Ethics Statement
All experimental procedures with animals in present study were performed in accordance with the National Research Council’s Guide for the Care and Use of Laboratory Animals. The animal care and experimental protocols were approved by the Ethics Committee of Anhui University of Science and Technology, China (IEC No: 201902). The procedures for maintenance and treatment of laboratory animals were approved by the Animal Care and Use Committee of the Anhui University of Science and Technology.
Acknowledgments
Financial support was provided by Anhui Provincial Natural Science Foundation (2008085MH261, 2208085QH282), Anhui Medical University Research Foundation, China (2022xkj123), Scientific Research Project of Education Department of Anhui Province (2022AH052333).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Tasneem U, Mehmood K, Majid M, et al Methicillin resistant staphylococcus aureus: a brief review of virulence and resistance. J Pak Med Assoc. 2022;72(3):509–515. doi:10.47391/JPMA.0504
2. Lakhundi S, Zhang K. Methicillin-resistant staphylococcus aureus: molecular characterization, evolution, and epidemiology. Clin Microbiol Revs. 2018;31(4):e00020–18. doi:10.1128/CMR.00020-18
3. Rehman S. A parallel and silent emerging pandemic: antimicrobial resistance (AMR) amid COVID-19 pandemic. J Infect Public Health. 2023;16(4):611–617. doi:10.1016/j.jiph.2023.02.021
4. Alkharsah KR, Rehman S, Alkhamis F, et al Comparative and molecular analysis of MRSA isolates from infection sites and carrier colonization sites. Ann Clin Microbiol Antimicrob. 2018;17(1):7. doi:10.1186/s12941-018-0260-2
5. Güven Gökmen T, Kalayci Y, Yaman A, et al Molecular characterization of methicillin-resistant Staphylococcus aureus strains by spa typing and pulsed field gel electrophoresis methods. BMC Microbiol. 2018;18(1):155. doi:10.1186/s12866-018-1305-6
6. Vollmer W, Blanot D, de Pedro MA. Peptidoglycan structure and architecture. FEMS Microbiol Rev. 2008;32(2):149–167. doi:10.1111/j.1574-6976.2007.00094.x
7. Sutton JAF, Carnell OT, Lafage L, et al Staphylococcus aureus cell wall structure and dynamics during host-pathogen interaction. PLoS Pathog. 2021;17(3):e1009468. doi:10.1371/journal.ppat.1009468
8. Apostolos AJ, Pidgeon SE, Pires MM. Remodeling of cross-bridges controls peptidoglycan cross-linking levels in bacterial cell walls. Chem Biol. 2020;15(5):1261–1267. doi:10.1021/acschembio.0c00002
9. Pazos M, Peters K. Peptidoglycan. Subcell Biochem. 2019;92:127–168. doi:10.1007/978-3-030-18768-2_5
10. Shaku M, Ealand C, Matlhabe O, et al Peptidoglycan biosynthesis and remodeling revisited. Adv Appl Microbiol. 2020;112:67–103. doi:10.1016/bs.aambs.2020.04.001
11. Nagarajan R. Antibacterial activities and modes of action of vancomycin and related glycopeptides. Antimicrob Agents Chemother. 1991;35(4):605–609. doi:10.1128/AAC.35.4.605
12. Rodríguez-Tébar A, Vázquez D. Penicillin-binding proteins and peptidoglycan peptide-interacting proteins. Microbiol Sci. 1984;1(9):211–214.
13. Wang M, Buist G, van Dijl JM. Staphylococcus aureus cell wall maintenance - The multifaceted roles of peptidoglycan hydrolases in bacterial growth, fitness, and virulence. FEMS. Microbiol Rev. 2022;46(5):fuac025. doi:10.1093/femsre/fuac025
14. Rohs PDA, Bernhardt TG. Growth and division of the peptidoglycan matrix. Rev Microbiol. 2021;75:315–336. doi:10.1146/annurev-micro-020518-120056
15. Dubrac S, Boneca IG, Poupel O, Msadek T. New insights into the WalK/WalR (YycG/YycF) essential signal transduction pathway reveal a major role in controlling cell wall metabolism and biofilm formation in Staphylococcus aureus. J Bacteriol. 2007;189(22):8257–8269. doi:10.1128/JB.00645-07
16. Buchad H, Nair M. The small RNA sprx regulates the autolysin regulator walr in Staphylococcus aureus. Microbiol Res. 2021;250:126785. doi:10.1016/j.micres.2021.126785
17. Kathirvel M, Buchad H, Nair M. Enhancement of the pathogenicity of Staphylococcus aureus strain Newman by a small noncoding RNA sprx1. Med Microbiol Immunol. 2016;205(6):563–574. doi:10.1007/s00430-016-0467-9
18. Nega M, Tribelli PM, Hipp K, et al New insights in the coordinated amidase and glucosaminidase activity of the major autolysin (Atl) in Staphylococcus aureus. Commun Biol. 2020;3(1):695. doi:10.1038/s42003-020-01405-2
19. Bose JL, Lehman MK, Fey PD, Bayles KW. Contribution of the Staphylococcus Aureus Atl AM and GL murein hydrolase activities in cell division, autolysis, and biofilm formation. PLoS One. 2012;7(7):e42244. doi:10.1371/journal.pone.0042244
20. Houston P, Rowe SE, Pozzi C, et al Essential role for the major autolysin in the fibronectin-binding protein-mediated Staphylococcus aureus biofilm phenotype. Infect Immun. 2011;79(3):1153–1165. doi:10.1128/IAI.00364-10
21. Xie Y, Wang L, Yang Y, et al Antibacterial and anti-biofilm activity of diarylureas against Enterococcus faecium by suppressing the gene expression of peptidoglycan hydrolases and adherence. Front Microbiol. 2022;13:1071255. doi:10.3389/fmicb.2022.1071255
22. Brückner R. Gene replacement in Staphylococcus carnosus and Staphylococcus xylosus. FEMS Microbiol Lett. 1997;151(1):1–8. doi:10.1111/j.1574-6968.1997.tb10387.x
23. Biswas R, Voggu L, Simon UK, et al Activity of the major staphylococcal autolysin Atl. FEMS Microbiol Lett. 2006;259(2):260–268. doi:10.1111/j.1574-6968.2006.00281.x
24. Chen Q, Zhang Q, Cao P, et al NOD2-mediated HDAC6/NF-κb signalling pathway regulates ferroptosis induced by extracellular histone H3 in acute liver failure. J Cell Mol Med. 2022;26(21):5528–5538. doi:10.1111/jcmm.17582
25. Zhu QY, Tai S, Tang L, et al N-acetyl cysteine ameliorates aortic fibrosis by promoting M2 macrophage polarization in aging mice. Redox Rep. 2021;26(1):170–175. doi:10.1080/13510002.2021.1976568
26. Gao K, Su B, Dai J, et al Anti-Biofilm and Anti-Hemolysis Activities of 10-Hydroxy-2-decenoic Acid against Staphylococcus aureus. Molecules. 2022;27(5):1485. doi:10.3390/molecules27051485
27. Günther G, Herlax V, Lillo MP, et al Study of rabbit erythrocytes membrane solubilization by sucrose monomyristate using laurdan and phasor analysis. Colloids Surf B Biointerfaces. 2018;161:375–385. doi:10.1016/j.colsurfb.2017.10.068
28. Ouyang J, Sun F, Feng W, et al Antimicrobial activity of galangin and its effects on murein hydrolases of vancomycin-intermediate Staphylococcus aureus (VISA) strain mu50. Chemotherapy. 2018;63(1):20–28. doi:10.1159/000481658
29. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi:10.1006/meth.2001.1262
30. Wang M, van den Berg S, Mora Hernández Y, et al Differential binding of human and murine IgGs to catalytic and cell wall binding domains of Staphylococcus aureus peptidoglycan hydrolases. Sci Rep. 2021;11(1):13865. doi:10.1038/s41598-021-93359-6
31. Salamaga B, Kong L, Pasquina-Lemonche L, et al Demonstration of the role of cell wall homeostasis in Staphylococcus aureus growth and the action of bactericidal antibiotics. Proc Natl Acad Sci U S A. 2021;118(44):e2106022118. doi:10.1073/pnas.2106022118
32. Liu G, Xiang H, Tang X, et al Transcriptional and functional analysis shows sodium houttuyfonate-mediated inhibition of autolysis in Staphylococcus aureus. Molecules. 2011;16(10):8848–8865. doi:10.3390/molecules16108848
33. Ledala N, Wilkinson BJ, Jayaswal RK. Effects of oxacillin and tetracycline on autolysis, autolysin processing and atl transcription in Staphylococcus aureus. Int J Antimicrob Agents. 2006;27(6):518–524. doi:10.1016/j.ijantimicag.2006.03.008
34. Antignac A, Sieradzki K, Tomasz A. Perturbation of cell wall synthesis suppresses autolysis in Staphylococcus aureus: evidence for coregulation of cell wall synthetic and hydrolytic enzymes. J Bacteriol. 2007;189(21):7573–7580. doi:10.1128/JB.01048-07
35. Tomasz A, Albino A, Zanati E. Multiple antibiotic resistance in a bacterium with suppressed autolytic system. Nature. 1970;227:5254):138–140. doi:10.1038/227138a0
36. Chan YG, Frankel MB, Missiakas D, et al SagB glucosaminidase is a determinant of staphylococcus aureus glycan chain length, antibiotic susceptibility, and protein secretion. J Bacteriol. 2016;198(7):1123–1136. doi:10.1128/JB.00983-15
37. Bersch KL, DeMeester KE, Zagani R, et al Bacterial peptidoglycan fragments differentially regulate innate immune signaling. ACS Cent Sci. 2021;7(4):688–696. doi:10.1021/acscentsci.1c00200
38. Brahma B, Patra MC, Mishra P, et al Computational studies on receptor-ligand interactions between novel buffalo (Bubalus bubalis) nucleotide-binding oligomerization domain-containing protein 2 (NOD2) variants and muramyl dipeptide (MDP). J Mol Graph Model. 2016;65:15–26. doi:10.1016/j.jmgm.2016.02.004
39. Gao J, Wang L, Jiang J, et al A probiotic bi-functional peptidoglycan hydrolase sheds NOD2 ligands to regulate gut homeostasis in female mice. Nat Commun. 2023;14(1):3338. doi:10.1038/s41467-023-38950-3
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.





