Back to Journals » International Journal of Nanomedicine » Volume 13
In vivo antimicrobial activity of silver nanoparticles produced via a green chemistry synthesis using Acacia rigidula as a reducing and capping agent
Authors Escárcega-González CE , Garza-Cervantes JA , Vazquez-Rodríguez A , Montelongo-Peralta LZ , Treviño-Gonzalez MT , Díaz Barriga-Castro E. , Saucedo-Salazar EM, Chávez Morales RM , Regalado-Soto DI , Treviño-González FM , Carrazco Rosales JL , Villalobos Cruz R , Morones-Ramirez JR
Received 22 December 2017
Accepted for publication 24 February 2018
Published 17 April 2018 Volume 2018:13 Pages 2349—2363
DOI https://doi.org/10.2147/IJN.S160605
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Thomas Webster
Carlos Enrique Escárcega-González,1,2 JA Garza-Cervantes,1,2 A Vázquez-Rodríguez,1,2 Liliana Zulem Montelongo-Peralta,1,2 MT Treviño-González,3 E Díaz Barriga Castro,4 EM Saucedo-Salazar,4 RM Chávez Morales,5 DI Regalado Soto,5 FM Treviño González,5 JL Carrazco Rosales,6 Rocío Villalobos Cruz,6 José Rubén Morones-Ramírez1,2
1Universidad Autónoma de Nuevo León, Facultad de Ciencias Químicas, Pedro de Alba, S/N, San Nicolás de los Garza, Nuevo León, México; 2Centro de Investigación en Biotecnología y Nanotecnología, Facultad de Ciencias Químicas, Universidad Autónoma de Nuevo León, Parque de Investigación e Innovación Tecnológica, Apodaca, Nuevo León, México; 3Universidad Autónoma de Nuevo León, Facultad de Ingeniería Mecánica y Eléctrica, Pedro de Alba, S/N, San Nicolás de los Garza, Nuevo León, México; 4Centro de Investigación en Química Aplicada, Saltillo, Coah, México; 5Departamento de Fisiología y Farmacología, Centro de Ciencias Básicas, Universidad Autónoma de Aguascalientes, Colonia Ciudad Universitaria, Aguascalientes, México; 6Departamento de Microbiología, Centro de Ciencias Básicas, Universidad Autónoma de Aguascalientes, Colonia Ciudad Universitaria, Aguascalientes, Mexico
Introduction: One of the main issues in the medical field and clinical practice is the development of novel and effective treatments against infections caused by antibiotic-resistant bacteria. One avenue that has been approached to develop effective antimicrobials is the use of silver nanoparticles (Ag-NPs), since they have been found to exhibit an efficient and wide spectrum of antimicrobial properties. Among the main drawbacks of using Ag-NPs are their potential cytotoxicity against eukaryotic cells and the latent environmental toxicity of their synthesis methods. Therefore, diverse green synthesis methods, which involve the use of environmentally friendly plant extracts as reductive and capping agents, have become attractive to synthesize Ag-NPs that exhibit antimicrobial effects against resistant bacteria at concentrations below toxicity thresholds for eukaryotic cells.
Purpose: In this study, we report a green one-pot synthesis method that uses Acacia rigidula extract as a reducing and capping agent, to produce Ag-NPs with applications as therapeutic agents to treat infections in vivo.
Materials and methods: The Ag-NPs were characterized using transmission electron microscopy (TEM), high-resolution TEM, selected area electron diffraction, energy-dispersive spectroscopy, ultraviolet–visible, and Fourier transform infrared.
Results: We show that Ag-NPs are spherical with a narrow size distribution. The Ag-NPs show antimicrobial activities in vitro against Gram-negative (Escherichia coli, Pseudomonas aeruginosa, and a clinical multidrug-resistant strain of P. aeruginosa) and Gram-positive (Bacillus subtilis) bacteria. Moreover, antimicrobial effects of the Ag-NPs, against a resistant P. aeruginosa clinical strain, were tested in a murine skin infection model. The results demonstrate that the Ag-NPs reported in this work are capable of eradicating pathogenic resistant bacteria in an infection in vivo. In addition, skin, liver, and kidney damage profiles were monitored in the murine infection model, and the results demonstrate that Ag-NPs can be used safely as therapeutic agents in animal models.
Conclusion: Together, these results suggest the potential use of Ag-NPs, synthesized by green chemistry methods, as therapeutic agents against infections caused by resistant and nonresistant strains.
Keywords: silver nanoparticles, green synthesis, in vitro antibacterial activity, in vivo antibacterial activity, skin infection, toxicological study
Introduction
Over the past few decades, there has been a growing interest in exploring alternative strategies to design and develop new antimicrobials due to the increased incidence in microbial resistance against commonly used antibiotics.1,2 The field of nanotechnology offers a toolbox of synthesis and design strategies to develop antimicrobial nanotherapeutics with tailored antimicrobial properties to counterattack the problem of antimicrobial resistance. Several reports have shown that nanoparticles made up of different noble metals can be used to kill both resistant and nonresistant bacterial strains.87–89 The large surface area-to-volume ratio exhibited by these systems at the nanoscale has been hypothesized to be one of the bases of the antimicrobial effects against a broad spectrum of microorganisms.1 Silver nanoparticles (Ag-NPs) have obtained considerable attention due to their attractive physicochemical properties2,3 and remarkable antimicrobial effects exhibited against Gram-negative and Gram-positive multidrug-resistant bacteria.4 The bactericidal effect of silver in its various chemical forms has been exploited since ancient civilizations;5 during the 18th century, silver was commonly used to treat infected ulcers, and more recently, in the early 1920s, Ag-NPs were included for wound treatment by the US Food and Drug Administration6 due to their strong toxicity against a wide range of microorganisms.7–9 There are various mechanisms that have been linked to the toxicity of silver compounds against microorganisms, among the most common are direct disruptive interactions with the cell membrane and DNA2 and the trigger of biochemical cascades that lead to free radical production, altering cell membrane permeability.10 In the case of Ag-NPs, it has been shown that there is a correlation between size and the exhibited antimicrobial effect.2 Nevertheless, the use of Ag-NPs for antimicrobial therapies has two major drawbacks: the toxicity exhibited to eukaryotic and mammalian systems and the toxicity of the chemical synthesis to produce them. Therefore, there have been efforts toward the development of greener synthesis processes to produce Ag-NPs11,12 with tailored properties that reduce toxicity against eukaryotic systems.13 Although there are various methods to synthesize Ag-NPs, including chemical reduction,14 photochemical reduction,15,16 electrochemical reduction,17 vaporization, and thermal synthesis,18 there is a growing interest in developing a green synthesis method to produce nanoparticles using biological systems19,20 and plant extracts.21 Notwithstanding the diversity of green synthesis approaches, the use of plant extracts, as reductive media for nanoparticle synthesis, is one of the most convenient routes.22,23
Ag-NPs have been synthesized using extracts obtained from different plants such as Acorus calamus (rhizome), Boerhaavia diffusa (whole plant), tea extract (leaves), Tribulus terrestris (fruit), Aloe vera (leaves), Eucalyptus hybrid (peel), Memecylon edule (leaves), and Citrus sinensis (peel).24 In fact, within the last decade, hundreds of plant extracts have been reported to be effective reducing agents in the synthesis of Ag-NPs.25 These methods have been widely explored since they provide environmentally friendly and cost-effective routes to the synthesis of metallic nanostructures, in addition to being easily scalable production processes.26–28 In this study, we developed a green synthesis method to produce Ag-NPs using the extract of an Acacia rigidula plant. This plant is commonly known as blackbrush acacia, and it is a shrub that can be found in the regions of west and southwest Texas, as well as the Mexican northeastern region.29 Although there is limited information on the chemical composition of the A. rigidula plant,30 gums and condensed tannins are the most characterized parts in the literature.31 Forty-four amines and alkaloids,29 such as phenethylamines, have been identified in A. rigidula. These components have been associated with promotion of weight loss; therefore, both the A. rigidula plant and its extracts have been extensively used as dietary supplements.30 Furthermore, forages from A. rigidula have been widely explored as a protein supplement to feed cattle.32,33 It was then interesting to explore, in this study, the potential use of the extract, obtained from waste parts of the A. rigidula plant (roots and stems), as a reducing and stabilizing agent in the synthesis of Ag-NPs. The successfully obtained Ag-NPs showed antimicrobial activity in vitro against Gram-negative bacteria (Escherichia coli, Pseudomonas aeruginosa, and a multidrug-resistant P. aeruginosa strain) and Gram-positive bacteria (Bacillus subtilis). Moreover, in vivo studies showed promising antimicrobial effects of Ag-NPs in a murine skin infection model (using a P. aeruginosa clinical resistant strain). In addition, an in vivo toxicological study was carried out to verify the safety of using Ag-NPs; the results demonstrate them to be a promising treatment for skin infections in the clinic. The combined results presented in this work contribute toward the development and synthesis of alternative antimicrobial agents, nanotherapeutics, to treat infections caused by clinical multidrug-resistant strains.
Materials and methods
Biological materials
Stems and roots from the A. rigidula plant were collected in Nuevo León, Mexico. Gram-negative bacteria used in the antimicrobial assays were E. coli American Type Culture Collection (ATCC)-11229, P. aeruginosa ATCC 27853, and a clinical multidrug-resistant P. aeruginosa isolated at the San Vicente Hospital in Monterrey, Nuevo León, Mexico. The Gram-positive bacterium used in the antimicrobial assays was B. subtilis ATCC 23857.
Bacteria cultures were grown for 20 h at 37°C and 150 rpm (Lab Companion IS-971). After incubation, cultures were diluted 1:250 in fresh medium until an optical density (OD600) of 0.2±0.02 was reached, adjusting with fresh medium if necessary to reach a cell concentration range between 107 and 108 cells/mL. The resulting suspension was then diluted (1:100) with fresh medium, obtaining the bacterial working solution. Absorbance was measured using an ultraviolet–visible (UV–Vis) spectrophotometer (OPTIZEN 2120 UV plus), and cell number was monitored by the plate-counting method, measured by serial dilutions.
Synthesis of Ag-NPs
A. rigidula extract preparation
To prepare the extract, 5 g of stems and roots from the A. rigidula plant were mixed in 50 mL of distilled water. The mixture was then heated in a hotplate (Lab Companion HP-3100), until boiling, for 20 min. During the extraction time, agitation was performed using a magnetic stirrer. Once the infusion cooled, the extract was transferred to a 50-mL corning tube and centrifuged (Sorvall™ Leyend™ XFR; Thermo Fisher Scientific, Waltham, MA, USA) at 9,000 rpm for 10 min. Then, the supernatant was separated and vacuum-filtered (WP6111560; Merck Millipore, Billerica, MA, USA) using filter paper (Ahlstrom, Stockholm, Sweden). The obtained filtrate was stored at 4°C (Nor-Lake Scientific, Hudson, WI, USA).
Ag-NPs preparation
A total of 10 mL of AgNO3 0.1 M and 3 mL of A. rigidula extract were mixed in a 50 mL corning tube and heated in a hotplate for 1 h at 60°C. The formation of the Ag-NPs was monitored and determined through a change of appearance (color) of the mix that changed from transparent to brownish. The resulting solution with the Ag-NPs was centrifuged at 9,000 rpm for 15 min. Then, the product was washed with ethanol and was stored at −75°C for 24 h in an ultralow temperature freezer (Sanyo MDF-U53VA). Then, the product was lyophilized (FreeZone 6; Labconco, Kansas City, MO, USA) for 24 h to obtain the purified Ag-NPs.
Ag-NPs stability experiments
UV–Vis spectra (wavelength range from 200 to 800 nm) of the A. rigidula extract and the Ag-NPs (solution of 1 mg/mL in distilled water as a dispersive medium) were measured using a Multiskan G0 UV–Vis spectrophotometer (Thermo Fisher Scientific). To test stability of the synthesized Ag-NPs, UV–Vis spectra were obtained from Ag-NPs’ solution at two different time points (first time point right after being synthesized and second time point after being stored in solution for 4 months).
Characterization techniques
Morphological and structural characteristics of the Ag-NPs were studied with conventional transmission electron microscopy (TEM) and high-resolution TEM (HRTEM) and selected area electron diffraction (SAED) using an FEI-TITAN 80–300 kV microscope operated at an accelerating voltage of 300 kV. The specimen for these studies was prepared by depositing and evaporating a drop of the colloidal solution (Ag-NPs 1 mg/mL), previously dispersed by means of an ultrasonic cleaner (Bransonic® Branson 2510MT; Bransonic, Dansbury, CT, USA), onto lacey carbon-coated copper grids. The elements present in the samples were determined using an energy-dispersive spectrometry (EDS) analyzer integrated in the transmission electron microscope. In relation to the particle size, TEM images were used to measure the diameter of Ag-NPs to estimate the size distribution of our nanoparticles using the image processing software ImageJ – National Institute of Health (NIH). The images obtained from the SAED patterns were interpreted with simulated patterns using the Web-based Electron Microscopy Application Software (Web-EMAPS).34
Furthermore, UV–Vis spectra (wavelength range from 200 to 1,000 nm) of the Ag-NPs (solution of 1 mg/mL in distilled water as a dispersive medium) were obtained using a Multiskan G0 UV–Vis spectrophotometer. To obtain information related to the interaction between A. rigidula extract and Ag+ ions to produce the Ag-NPs and their capping, infrared spectra were obtained for the extract alone and for the Ag-NPs. The spectra were acquired using a Fourier transform infrared (FT-IR) spectrophotometer (Nicolet™ iS™ 50; Thermo Fisher Scientific). Samples from the extract solution and from a 1 mg/mL sample of Ag-NPs were freeze-dried with a lyophilizer (FreeZone 6) for 24 h to obtain the samples, which were diluted in KBr, using the attenuated total reflectance (ATR) method.
In vitro antibacterial test
Minimum inhibitory concentrations (MICs)
MIC assays were assessed in 96-well plates (Costar; Corning Incorporated, Corning, NY, USA) based on a modified methodology reported previously35 and National Committee for Clinical Laboratory Standards.36 Stocks of the Ag-NPs were prepared at a final concentration of 1 mg/mL using distilled water as the vehicle. To perform the MIC tests, Ag-NPs’ stock solution was added to reach a 0.5 mg/mL concentration within a final volume of 200 μL. Next, serial dilutions of the Ag-NPs were performed by taking 100 μL from the previous solution to every next well, with 100 μL of culture media, and discarding the last 100 μL from the last dilution so the tested concentrations were 0.25–4.88×10−4 mg/mL for Ag-NPs. Then, 100 μL of the bacteria working solution was added to each test well to achieve a final concentration of 105 cells/mL, followed by incubation at 37°C and 150 rpm. After 20 h of incubation at 37°C, the MIC was measured. The MIC for Ag-NPs was defined as the concentration at which no significant growth was observed by the naked eye. All tests, and their respective control samples, were performed in triplicate.
Minimum bactericidal concentration (MBC)
To assess the bactericidal activity of the synthesized Ag-NPs, 1, 2, and 4 times, the Ag-NPs’ MIC was tested against the bacterial strains. In a typical assay, an overnight culture of each strain was diluted 1:250 in 5 mL of fresh Luria Broth (LB) medium, followed by incubation at 37°C and 150 rpm, until it reached exponential phase at a critical OD (OD600 =0.18–0.22). A total of 20 μL of the exponential cultures were transferred to 96-well plates, which contained the Ag-NP treatments, to be incubated at 37°C and 150 rpm for 20 h. After incubation, the colony-forming units (CFUs) per milliliter were determined through the drop count method. Tenfold diluted aliquots were obtained, and then, three 10 μL drops of each of the diluted samples were placed on LB agar plates. The plates were then incubated at 37°C for 24 h, and viable bacterial colonies were counted. All tests and their respective control samples were performed in replicates of three.
Fluorescence microscopy imaging of treated and untreated bacteria
Fluorescence microscopy of the treated and untreated bacteria was carried out based on the methodology described by Vazquez-Rodriguez et al.37 A total of 105 bacterial cells of the resistant P. aeruginosa clinical strain were treated with Ag-NPs, at 0, 0.5, 1, and 2 times the MIC, for 20 h at 37°C and 150 rpm. After the 20 h treatment, bacterial cells were stained with 5 μg/mL of propidium iodide (PI) for 1 h at 37°C and 150 rpm. A total of 20 μL of the treated samples and the control were placed and spread in glass slides, previously cleaned with 96% ethanol, allowing the samples to air dry and fix by adding 200 μL of methanol for 2 min. The methanol excess was decanted, and the slides were then washed twice with phosphate-buffered saline (PBS). Micrograph images were obtained under DM3000 (Leica Microsystems, Wetzlar, Germany) using the 100× objective with an Y3 ET filter system.
In vivo antibacterial test
Animals
Female adult Wistar rats (200–250 g) were used and maintained in stainless steel cages with a 12 h light/dark regime. All the experimental animals were handled ethically (refer the “Ethics approval and informed consent” section).
In vivo skin infection model
The in vivo experiment based on a tape stripping infection model38 was performed with some modifications. The rats were anesthetized with pentobarbital (Pentosedal; Laboratorios Maver, Ciudad de México, Mexico) injecting intraperitoneally 2.5 mL/kg of body weight (bw). Once anesthetized, the back of the rats was shaved in an area of 3 cm2 with a trimmer (274 Series A; Oster, Boca Raton, FL, USA). Next, an area of 2.5 cm2 of the hairless skin of the rats was tape stripped five times in succession using an elastic adhesive bandage (Sedasiva®; BSN Medical, Yumbo, Colombia). Damage on the skin was defined as the reddening and glistening of the skin with the absence of regular bleeding. This procedure was performed in all the animals used in the experiment. After that, to cause infection, bacterial inoculation was performed by placing, in the wound produced with the tape stripping process, 100 μL of a 107 cells/mL P. aeruginosa culture in log phase. The P. aeruginosa culture was obtained from an overnight culture grown as described previously in the MBC description of the Materials and methods section.
Experimental design
The rats were divided into three groups (three animals per experimental group [n=3]): the control group (with the wound on the skin without infection, and treated with 100 μL of saline solution every 24 h for 3 days in the damaged zone), the infected group (treated in the damaged zone with 100 μL of the clinical strain of P. aeruginosa culture), and the Ag-NP-treated group (treated with 100 μL of Ag-NPs solution at 200 parts per million [ppm] every 24 h for 3 days after infection with the clinical strain of P. aeruginosa in the damaged zone). During all the experiments, the rats were kept with food and water ad libitum at room temperature (24°C±1°C).
The rats were anesthetized and sacrificed 24 h after the third day of treatment, and 300 mg of the wound (~1 cm2) was removed and homogenized (Tissue-Tearor 398; BioSpec, Bartlesville, OK, USA) with 3 mL of phosphate buffer (100 mgwound/mLbuffer).
Finally, 10 μL of homogenates (to have a 1:100 dilution) were used to carry out a pour plate technique to determine the CFUs of living bacteria and to compare these results among the study groups.
In vivo toxicological study
Experimental design
To demonstrate the safety in the use of the Ag-NPs produced in this study, four groups of female adult Wistar rats (200–250 g) with three animals per group (n=3) were used to carry out the toxicological test. The four groups included the control group and three groups treated with the Ag-NPs at different concentrations. In this experiment, the rats were injured based on the tape stripping infection model described previously in the in vivo skin infection model methodology. Briefly, the pentobarbital was administered intraperitoneally to the rats (2.5 mL/kg bw) to anesthesize them. After that, a 3 cm2 area from the back of the rats was shaved using a trimmer in which 2.5 cm2 of the shaved part was tape stripped five times with an elastic adhesive bandage until the hairless skin reddened and glistened, but there was no bleeding observed. This procedure was done in all of the animals used in this experiment. To start the test, the animals in the control group were treated with 100 μL of saline solution on the wound produced by the tape stripping procedure, while the rats in the other three tested groups were treated with 100, 200, and 400 ppm of Ag-NPs, respectively. The treatment consisted of applying either saline solution or the Ag-NPs solution (at the respective concentration) directly on the wound until the solution was absorbed through the skin. All groups were treated in the damaged zone every 24 h for 3 days; the same way as the antibacterial assay was performed. Throughout the experiment, the rats were kept with food and water ad libitum at room temperature (24°C±1°C). Then, 24 h after the third day of treatment, urine samples were collected in vessels attached with metabolic cages (3700M020; Tecniplast, Buguggiate (VA), Italy) in which the rats were placed. Afterward, with the rats previously anesthetized, blood samples were also obtained and the rats were sacrificed.
Biochemical assays
In order to test kidney and liver functions of the rats exposed to Ag-NPs, the urine volume was measured, and with their respective methods, the concentration of creatinine,39,40 glucose,41 and proteins42 in urine was also measured using a UV–Vis spectrophotometer (DMS 80; Varian, Palo Alto, CA, USA). For the case of blood plasma samples, the concentration of creatinine,39,40 albumin,43 alanine aminotransferase (ALT),44 and aspartate aminotransferase (AST)45 was measured using a UV–Vis spectrophotometer (DMS 80).
Likewise, during the experiment, the appearance of the skin (wound) of the experimental groups exposed to Ag-NPs was observed and compared with the control group.
Statistical analysis
For the in vivo study (antibacterial and toxicological experiments), the significance of the differences between group averages was calculated with the two-tailed Student’s t-test for grouped data with an analysis of variance (ANOVA) posttest. The calculations and graphs were prepared using the Prism 4 software (GraphPad Software, Inc., La Jolla, CA, USA).
Ethics approval and informed consent
Animal care and experimentation practices at the Universidad Autónoma de Aguascalientes are constantly evaluated by the Animal Care and Use Committee, adhering to the official Mexican regulations (NOM-062-ZOO-1999). Mexican regulations are in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the NIH and The Weatherall Report of the USA to ensure compliance with established international regulations and guidelines. Experimental protocols were approved by the local Institutional Bioethical Committee. At the end of experiments, animals were sacrificed by using an excess of sodium pentobarbital anesthesia (40 mg/kg bw). Efforts were made to minimize suffering.
Results
Characterization of Ag-NPs
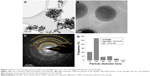
Ag-NPs were produced in an aqueous solution containing an extract from A. rigidula. As can be observed in the TEM and HRTEM micrographs (Figure 1A and B), the Ag-NPs display homogeneous spherical shapes (Figure 1A and B). An SAED pattern of the Ag-NPs showed spotty rings that can be attributed to the (111), (200), (220), (311), and (331) planes of a face-centered cubic (fcc) metallic Ag-NP (Figure 1C). The Ag-NPs exhibit a diameter size distribution that ranges from 8 to 66 nm with a mean of 22.46 nm and a standard deviation of 10.83 nm (Figure 1D). The histogram in Figure 1D shows that most of the Ag-NPs have a diameter size between 15 and 25 nm, which corresponds to a narrow homogeneous distribution.
Further characterization of the nanoparticles by EDS analysis shows the presence of Ag, which confirms the presence of Ag-NPs (Figure 2). In the EDS analysis, other elements such as C and Cu were detected, where C corresponds to the organic matrix and lacey carbon of the TEM grid and Cu is observed since the grid used for the TEM analysis is made up of Cu (Figure 2).
  | Figure 2 EDS spectrum of the Ag-NPs with the peaks labeled. |
The UV–Vis spectra of the A. rigidula alone, and the Ag-NPs produced by reduction and capping using the A. rigidula extract, are shown in Figure 3A. The Ag-NPs exhibit a plasmon resonance spectrum with an absorption band between 430 and 480 nm. Furthermore, Figure 3A shows a dotted UV–Vis spectra that corresponds to the Ag-NPs’ sample that was stored in solution for 4 months. As can be observed, the spectra present minimal changes and keep their absorption peak between 430 and 480 nm. This result demonstrates the stability of the Ag-NPs after being stored for long time periods. FT-IR characterization of the extract alone (Figure 3B) and the Ag-NPs synthesized with the extract (Figure 3C) showed that both samples contained functional groups that have been previously reported to be present in an A. rigidula extract.46–51 Some of the characteristic peaks observed in both samples include (Figure 3B and C) a prominent peak at 3,419 cm−1 due to the stretching of O–H in alcohols and phenols; two peaks at 2,929 and 2,854 cm−1 due to the stretching of CH3− and CH2− in aliphatic compounds; one peak at 1,614 cm−1 due to the bending of N−H in amines; one peak at 1,517 cm−1 corresponding to the stretching of C=C in aromatic compounds; another peak at 1,447 cm−1 attributed to −C−N groups; one peak at 1,384 cm−1 due to the symmetric deformation of CH3 in aromatic and aliphatic compounds; another peak at 1,285 cm−1 corresponding to the wagging of C−H in alkyl halides; a peak at 1,250 cm−1 due to the stretching of C=O groups; two peaks at 1,114 and 1,068 cm−1 due to the stretching of C−O in polysaccharides; three peaks at 894, 823, and 767 cm−1 due to the bending of =CH in aromatic hydrocarbons; and one peak at 689 cm−1 attributed to the bending of C−H in alkynes.
In vitro antimicrobial activity of Ag-NPs in Gram-negative and Gram-positive bacteria
MIC and MBC assays were performed under the same culture conditions for each of the bacteria tested in this work. B. subtilis had the lowest MIC with a value of 0.48 ppm. For P. aeruginosa, the obtained MIC was 15.6 ppm, twice the MIC obtained for the clinical multidrug resistant bacteria of the same strain. The MIC value obtained for E. coli was 62.5 ppm, the highest of all the MICs determined. The MIC values obtained in this experiment are summarized in Table 1.
  | Table 1 Nominal MICs of Ag-NPs for the four different strains tested in this work |
Bactericidal activity was tested through MBC assays using Ag-NPs at different treatment concentrations linked to the MICs reported. As can be observed in Figure 4A–D, E. coli and B. subtilis show the lowest MBC (two times the MIC). In addition, with a statistical significance (P<0.05), even at one MIC, the treatments show bactericidal activity. For P. aeruginosa strains, the MBCs were found at four times the MICs. However, as observed for E. coli and B. subtilis, bactericidal activity can be observed as low as the concentration corresponding to one MIC. Thus, the Ag-NPs produced by A. rigidula extract show bactericidal activity in both Gram-negative and Gram-positive bacteria.
Morphological changes in P. aeruginosa after treatment with Ag-NPs
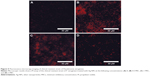
Since antimicrobial activity has been extensively linked to bacterial envelope damage, morphological changes were explored in a clinically relevant multidrug-resistant P. aeruginosa that was treated with Ag-NPs. Bacterial membrane disruption caused by the Ag-NPs treatments was tested through a fluorescence PI stain assay.10 As can be observed in Figure 5, when compared with the untreated bacteria (Figure 5A), the florescent micrographs (Figure 5B–D) show a general qualitative increased fluorescence for all the treatments (0.5, one, and two times the MIC). These results suggest that bacterial membrane integrity is compromised either directly or as a consequence of the antimicrobial activity exhibited by the Ag-NPs.
In vivo antimicrobial activity of Ag-NPs in a murine skin infection model triggered by a P. aeruginosa clinical strain
A murine skin infection model in rats was performed to test the ability of Ag-NPs to treat a topical infection caused by a resistant clinical strain (P. aeruginosa). As can be observed in Figure 6, there is a statistically significant higher amount of CFUs present in the group infected with the clinical strain of P. aeruginosa with respect to the control group (P<0.05). Moreover, the group infected with P. aeruginosa and treated with Ag-NPs exhibits a number of CFUs statistically similar to the control group and statistically much lower than the group infected with P. aeruginosa and not treated with the Ag-NPs (P<0.05).
In vivo toxicological assay of the Ag-NPs
A murine toxicity infection model in rats was performed to test the toxicity of Ag-NPs during treatment of a topical infection caused by a resistant clinical strain (P. aeruginosa). For the toxicity study, different parameters of renal function were evaluated. These parameters included urinary volume, urinary concentrations of proteins, glucose and creatinine and blood concentrations of creatinine. The results shown in Figure 7 demonstrate that the control urine volumes (Figure 7A), as well as glucose (Figure 7B), total proteins (Figure 7C), and creatinine (Figure 7D) concentration levels in the urine, are statistically similar (P<0.05) to the values obtained for all the treatments with different Ag-NP concentrations (100, 200, and 400 ppm). The same trend is observed for creatinine levels shown in the blood (Figure 7E). In addition, no statistically significant differences (P<0.05) were observed between the groups of rats treated with the Ag-NPs and the control groups in the parameters tested to evaluate hepatic function (albumin [Figure 8A], ALT [Figure 8B], and AST [Figure 8C] concentrations in blood plasma).
Finally, no differences were observed in the appearance and characteristics of the skin between the treated animals at any of the concentrations of Ag-NPs.
Discussion
Synthesis and characterization of Ag-NPs
It is well-known that plant crude extracts contain secondary metabolites, such as phenolic acids, flavonoids, alkaloids, and terpenoids, that can trigger redox reactions, such as those that take part in the synthesis of Ag-NPs, which are involved in the reduction in silver ions into metallic silver atoms that further assemble into nanostructures.52 Many species of the Acacia genus contain different types of secondary metabolites that can be used as reducing agents such as amines and alkaloids, nonprotein amino acids, cyanogenic glycosides, cyclitols, fatty acids and seed oils, fluoroacetate, gums, terpenes, diterpenes, phytosterols, triterpene genins and saponins, hydrolyzable tannins, condensed tannins, and flavonoids.31 However, the polysaccharides or gums and complex phenolic compounds or condensed tannins are among the most studied and most recognized.31 Thus, the effective reducing capability of the A. rigidula extract used in the synthesis of Ag-NPs can be attributed to the phenolic compounds, saponins, and flavonoids.
The homogeneity of the size distributions observed for the Ag-NPs reported in this work compares with other works in the literature that report Ag-NPs produced by green synthesis methods. Ag-NPs have been obtained with sizes between 50 and 250 nm using Carica papaya leaf extract;53 with average sizes 23±12, 32±24 and 19±22 nm using Illicium verum (star anise) seed extracts,54 and average size 73 nm using an aqueous extract of Tephrosia tinctoria.55 Regarding spherical shape homogeneity, our synthesis method was compared with that of other works where spherical Ag-NPs are synthesized, such as in the case of other green synthesis methods that employ leaf extracts of Saraca indica,56 Ficus carica,57 and Cocos nucifera.58
The chemical and physical characterization of the Ag-NP samples suggests that the Acacia extract is capable of not only reducing the silver ions but also capping the Ag-NPs and stabilizing their surface. This is a phenomenon not commonly observed, but it has been previously reported that when both reducing and capping effects are present in an extract, it allows a tighter control of the nanoparticle sizes obtained and prevents further aggregation of the samples in time.52,59
The well-defined spotty rings correspond to (111), (200), (220), (311), and (331) planes of fcc metallic silver (Joint Committee on Powder Diffraction Standards file no 87-0720), while EDS analysis demonstrates that the nanoparticles obtained are constituted of silver and makes evident their crystalline nature (Figures 1C and 2). Similar information from the EDS and SAED analyses of Ag-NPs has been reported when using I. verum (star anise) seeds54 and extracts of Capsicum annuum var. aviculare (piquin) fruits.12
The UV–Vis spectrum of both the A. rigidula extract and the Ag-NPs shows that for the extract, the absorbance was low, but in the case of Ag-NPs, it displays the maximum absorbance at 444 nm, indicating that the extinction band is a result of the surface plasmon resonance (SPR) phenomenon produced by Ag-NPs (Figure 3A). These results correlate with previous reports that show a maximum absorption peak at 480 nm, when Ag-NPs were synthesized using T. tinctoria extract as a reducing agent,55 and an absorption peak at 420 nm when Ag-NPs were produced using Chrysopogon zizanioides aqueous extract.59 Hence, these results agree with the Mie scattering theory about single spherical Ag-NPs well dispersed in water with different diameters.54 In this regard, the width of the peak obtained in the UV–Vis spectrum is mainly attributed to the size distribution and differences in the shape of the Ag-NPs.60 Furthermore, the UV–Vis spectra of a sample that was stored in solution for 4 months show a sustained maximum absorbance at ~450 nm (Figure 3A), strongly suggesting that Ag-NPs remain stable during considerable long periods of time. Thus, the organic matrix of A. rigidula extract acts as an effective stabilizing agent.
In the case of the FT-IR spectroscopy (Figure 3B and C), the different functional groups observed in the Ag-NPs spectrum also appear in the A. rigidula extract. In addition, the peaks observed are in accordance with the different components that the extract may contain, such as carbohydrates, flavonoids, and polyphenols, that can be carrying out the process of reduction in the Ag+ ions to produce Ag-NPs. Therefore, the characterization data strongly indicate the function of the extract as a capping and stabilizing agent of the Ag-NPs. Moreover, the results suggest the presence of the components of the extract in the surface of the Ag-NPs.
Antibacterial test: in vitro and in vivo study
For >2,000 years, silver has been recognized for its medicinal properties. Hence, silver has been used as an antimicrobial agent due to its effectiveness against some pathogenic microorganisms and its lower toxicity in animal cells in comparison with other metals.24 Moreover, recently, there has been an increasing interest in the application of silver as nanoparticles, for medical applications, as an alternative antimicrobial agent. Taking into account the problem of multidrug resistance to the conventional antibiotics, both Gram-negative and Gram-positive bacteria have developed recently.61 For this reason, there is a need to search and produce more efficient antimicrobial agents that can fight strains of multidrug-resistant bacteria.62
The Ag-NPs produced by green synthesis with A. rigidula extract exhibited antimicrobial activity against Gram-negative bacteria (E. coli, P. aeruginosa, and the clinical strain of P. aeruginosa) and Gram-positive bacteria (B. subtilis) tested in our study (Table 1 and Figure 4A–D). Interestingly, the MIC values observed in our study for P. aeruginosa are similar to the values obtained by Ag-NPs produced from an aqueous extract of Phyllanthus amarus against several P. aeruginosa clinical strains,63 where MIC values varied from 6.25 to 12.5 ppm. MIC values obtained in this study for B. subtilis are lower than those previously reported, where a 12.5 ppm MIC value was observed with Ag-NPs obtained from an aqueous rhizome extract of Coptis chinensis.64 However, the same study showed lower MIC values for E. coli, where an MIC value of 25 ppm was reported.64 A similar MBC of the green synthesized Ag-NPs against the bacteria tested in this study was found using plant extracts as reducing and stabilizing agents.65–67 In this regard, it is possible to infer that Ag-NPs produced by an A. rigidula extract display effective antibacterial activity as bacteriostatic and bactericidal agents against both Gram-positive and Gram-negative bacteria.
We hypothesized that Ag-NPs might be interacting with both membrane and cell wall components, impeding bacterial growth. The PI assay results obtained by fluorescence microscopy (Figure 5A–D) show an increased fluorescence in the bacteria treated with Ag-NPs compared to the untreated control sample. This indicates that the Ag-NPs induce bacterial cell permeability by the disruption of cell membrane integrity. Thus, this can be one potential antibacterial mechanism linked to the effective antimicrobial properties exhibited by Ag-NPs. In this context, alterations in permeability and membrane structure have been mentioned as one mechanism of antimicrobial activity of Ag-NPs.68 Previous reports have shown that cell membrane damage of P. aeruginosa can be induced by Ag-NPs produced by green synthesis methods.69
For the in vivo study, we confirmed that the Ag-NPs’ antibacterial activity involved a reduction of CFUs in the group treated with the Ag-NPs. In comparison, Ag-NPs elaborated by means of an aqueous extract of Caltropis procera fruit or leaves, administered to infant mice colonized with Vibrio cholerae or enterotoxic E. coli, significantly reduced from 75 to 100 times the colonization rates of these pathogenic bacteria.70
Furthermore, nanobiocomposites of Ag-NPs synthetized with leaf extract of Syzygium cumini into a matrix of cellulose nanocrystals from Dendrocalamus hamiltonii and Bambusa bambos leaves showed antibacterial activity and were effective in obtaining an accelerated wound healing on, in vivo, skin of male, Swiss albino mice.71 Moreover, flavonoid-loaded Ag-NPs produced by a Ricinus communis leaf extracted flavonoid mixture were efficient in curing a Staphylococcus aureus infection on a silkworm Bombyx mori larvae.72 Therefore, molecules such as flavonoids have been found to be able to coat Ag-NPs,73,74 a similar to what we have seen in our study, and their presence in the surface could be a factor involved in the antimicrobial activity of these nanoparticles.
The main difference between Gram-negative and Gram-positive bacteria is the thickness of the cell wall. Gram-negative bacteria contain a thin peptidoglycan, 2–3 nm thick, between the outer membrane and the cytoplasmic or inner membrane,75 while Gram-positive bacteria possess a thicker peptidoglycan layer of ~30 nm thick, without outer membrane.76 In the case of B. subtilis (Gram-positive), growth inhibition can be attributed to interactions between Ag-NPs embedded in the cell wall, causing membrane permeability due to structural changes in the cell wall.77 In contrast, on Gram-negative bacteria such as E. coli and P. aeruginosa, the outer membrane is composed mainly of lipids and proteins, which are the first structures encountered by the Ag-NPs. Therefore, Ag-NPs could have adhered to these structures, altering their properties and integrity-forming gaps within the membrane, increasing its permeability.6 It is also well known that the membrane contains large amounts of sulfur-containing proteins and their inactivation can play a role in the deterioration of the membrane produced by Ag-NPs.78,79 Therefore, these kinds of proteins may be another target of the Ag-NPs, inactivating enzymes and contributing to the deterioration of the membrane.78,79 It has been reported that the antimicrobial activity of colloidal Ag-NPs is influenced by different particle characteristics such as size, shape, and capping agent.24,52,80 In this context, small sized Ag-NPs (as the Ag-NPs obtained in this study) have been reported to present a better bactericidal activity due to a greater binding surface in comparison to larger Ag-NPs.2 Moreover, the capping of the Ag-NPs produced in this study contains O−H functional groups (Figure 3) from polyphenol compounds present in the organic matrix. This functional group may produce damage and toxicity in the membrane and cell wall of the bacteria through oxidative stress by free radicals and reactive oxygen species (ROS).81 However, Ag-NPs alone are capable of producing ROS causing a microbial growth inhibition.82
Another mechanism that explains the toxicity of Ag-NPs against some bacteria refers to the release of Ag+ ions from the Ag-NPs. In fact, it has been reported that the toxicity of Ag-NPs with a size of 20–80 nm is predominantly due to the release of Ag+ ions.2,6 Since the Ag-NPs synthesized in this study (22.46 nm) are within the size range reported, it is possible that Ag+ ions release may also explain the growth inhibition shown here. This means that Ag+ ions could interact with membrane or cell wall components or participate in the generation of ROS. Hence, this mechanism can cause changes and damage in the permeability and physiology of bacteria. In addition, it has been shown that the release of Ag+ ions is the main antibacterial mechanism of toxicity of Ag-NPs.83
In vivo toxicological study
In an organism, the suitable functioning of its organs is very important to achieve a healthy balance maintaining the composition of its internal environment. This is largely achieved with the correct functioning of organs such as the kidney and liver. In the kidneys, the basic structural and functional units are the nephrons. They carry out their function by the following three fundamental processes: glomerular filtration, tubular reabsorption, and tubular secretion producing the final excretion of substances in the urine.84 In relation to the renal function parameters in urine assessed in this study, the kidneys regulate the volume (amount of body fluid), the concentration of glucose (associated with tubular function in nephrons), the concentration of total proteins (linked with glomerular filtration and general function of nephrons), and the concentration of creatinine both in plasma and urine (related with glomerular filtration rate in nephrons) in the organism. All the parameters measured in the treated samples were very similar to the values of the control group (Figure 7A–D). Thus, the Ag-NPs produced in our study did not produce any adverse or toxic effects in the nephrons and renal function of the animals treated.
Similarly, the liver is responsible for numerous vital functions for the proper functioning of higher order organisms, where the metabolization of chemical substances and biliary excretion occur.84 Therefore, this work analyzed hepatic function parameters in blood plasma when rats were treated with the Ag-NPs. The concentration of ALT and AST (biomarker of hepatic cell necrosis) and the concentration of albumin (biochemical biomarker of damage and hepatic function) of the treated groups with Ag-NPs were similar to the values of control groups (Figure 8A–C). Thus, the Ag-NPs obtained in this study did not have any adverse or toxic effects in the hepatic function. These findings are in agreement with other results that show zero mortality, absence of abnormal reactions, and minimal effect on kidney and liver indices in male and female Sprague Dawley rats administered orally with 2,000 mg/kg bw of Ag-NPs produced by Sargassum siliquosum, indicating low toxicity of these nanoparticles.85 Moreover, in the same study, it was observed that a pretreatment with Ag-NPs prevented liver cell damage caused by paracetamol at 100 and 200 mg/kg bw dose. Thus, Ag-NPs obtained by green synthesis provide a cheaper nontoxic method that ensures effectiveness and safety in the application of these nanoparticles in different fields of study.86 Finally, in the skin directly exposed to Ag-NPs, no adverse effects or interactions between tissue and the nanoparticles were found, since there were no alterations or changes in the appearance and structure of the treated skin. Therefore, the Ag-NPs at the tested concentration are safe for use in topical medical applications as an alternative antimicrobial therapeutic.
Conclusion
The use of the extract, obtained from roots and stems of A. rigidula, as a reducing and stabilizing agent represents a suitable alternative to produce antimicrobial metal nanoparticles (green synthesis). The synthesis reported here provides many advantages over conventional synthesis methods, such as the use and production of less toxic reagent and products. This allows for greater care of the environment. Currently, in the field of pharmacology and toxicology, there is a great need for better nontoxic treatment options that are more effective and have better antimicrobial activity against infectious diseases caused by different types of bacteria, especially against multidrug-resistant clinical strains. The antimicrobial properties shown by the Ag-NPs reported here represent a great alternative to achieve this objective. The results reported in this work are relevant to the field since they represent a contribution toward the development of more efficient and nontoxic alternatives for pharmacological treatments, taking advantage of the tools developed by nanotechnology. Nevertheless, further research in this subject is required to elucidate completely the interaction mechanism between Ag-NPs and bacteria. Moreover, research in pharmacological and toxicological studies, especially in vivo, is much needed to develop and design future antimicrobial therapeutic agents.
Availability of data and material
All data generated or analyzed during this study are included in this published article.
Acknowledgments
The authors thank the Universidad Autónoma de Nuevo León and CONACyT for financially supporting this work and Silvia Torres and JA Mercado Silva, who carried out some of the characterization studies reported herein, for providing technical support. This study was supported by Paicyt 2016–2017 Science Grant from the Universidad Autónoma de Nuevo León and by CONACyT Grants for Basic science (grant 221332), Fronteras de la Ciencia (grant 1502), and Infraestructura (grant 279957).
Author contributions
CEE-G, JAG-C, AV-R, LZM-P, and JRM-R designed, performed, and analyzed the experimental data and wrote the article. EDBC and EMS-S designed and performed the TEM analysis and contributed to the discussion and format of the article. RMCM, DIRS, FMTG, JLCR, and VCR designed and performed the antimicrobial experiments in vivo. MTT-G helped with the format and methodology of the article. All authors contributed toward data analysis, drafting and revising the paper and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
Khalil KA, Fouad H, Elsarnagawy T, Almajhdi FN. Preparation and characterization of electrospun PLGA/silver composite nanofibers for biomedical applications. Int J Electrochem Sci. 2013;8:3483–3493. | ||
Morones JR, Elechiguerra JL, Camacho A, et al. The bactericidal effect of silver nanoparticles. Nanotechnology. 2005;16(10):2346. | ||
Elechiguerra JL, Burt JL, Morones JR, et al. Interaction of silver nanoparticles with HIV-1. J Nanobiotechnology. 2005;3(1):6. | ||
Jha D, Thiruveedula PK, Pathak R, et al. Multifunctional biosynthesized silver nanoparticles exhibiting excellent antimicrobial potential against multi-drug resistant microbes along with remarkable anti-cancerous properties. Mater Sci Eng C Mater Biol Appl. 2017;80:659–669. | ||
Morones-Ramirez JR. Plata, metal precioso con amplio espectro de aplicaciones. Rev Cien Desarrollo. 2010;36(241):56–62. | ||
Durán N, Durán M, de Jesus MB, Seabra AB, Fávaro WJ, Nakazato G. Silver nanoparticles: a new view on mechanistic aspects on antimicrobial activity. Nanomedicine. 2016;12(3):789–799. | ||
Liau S, Read D, Pugh W, Furr J, Russell A. Interaction of silver nitrate with readily identifiable groups: relationship to the antibacterialaction of silver ions. Lett Appl Microbiol. 1997;25(4):279–283. | ||
Gupta A, Silver S. Molecular genetics: silver as a biocide: will resistance become a problem? Nat Biotechnol. 1998;16(10):888. | ||
Nomiya K, Yoshizawa A, Tsukagoshi K, Kasuga NC, Hirakawa S, Watanabe J. Synthesis and structural characterization of silver (I), aluminium (III) and cobalt (II) complexes with 4-isopropyltropolone (hinokitiol) showing noteworthy biological activities. Action of silver (I)-oxygen bonding complexes on the antimicrobial activities. J Inorg Biochem. 2004;98(1):46–60. | ||
Morones-Ramirez JR, Winkler JA, Spina CS, Collins JJ. Silver enhances antibiotic activity against Gram-negative bacteria. Sci Transl Med. 2013;5(190):190ra81. | ||
Mendoza-Reséndez R, Gómez-Treviño A, Barriga-Castro ED, Núñez NO, Luna C. Synthesis of antibacterial silver-based nanodisks and dendritic structures mediated by royal jelly. RSC Adv. 2014;4(4):1650–1658. | ||
Mendoza-Reséndez R, Núnez NO, Barriga-Castro ED, Luna C. Synthesis of metallic silver nanoparticles and silver organometallic nanodisks mediated by extracts of Capsicum annuum var. aviculare (piquin) fruits. RSC Adv. 2013;3(43):20765–20771. | ||
Geethalakshmi R, Sarada D. Gold and silver nanoparticles from Trianthema decandra: synthesis, characterization, and antimicrobial properties. Int J Nanomedicine. 2012;7:5375. | ||
Wang Y, Liu W, Liu W, et al. Synthesis of SnAgCu nanoparticles with low melting point by the chemical reduction method. Microelectron Reliabil. 2017;78:17–24. | ||
Gabriel JS, Gonzaga VA, Poli AL, Schmitt CC. Photochemical synthesis of silver nanoparticles on chitosans/montmorillonite nanocomposite films and antibacterial activity. Carbohydr Polym. 2017;171:202–210. | ||
dos Santos PL, Katic V, Toledo KC, Bonacin JA. Photochemical one-pot synthesis of reduced graphene oxide/Prussian blue nanocomposite for simultaneous electrochemical detection of ascorbic acid, dopamine, and uric acid. Sens Actuat B Chem. 2018;255(3):2437–2447. | ||
Winiarski JP, de Barros MR, Magosso HA, Jost CL. Electrochemical reduction of sulfite based on gold nanoparticles/silsesquioxane-modified electrode. Electrochim Acta. 2017;251:522–531. | ||
Landge S, Ghosh D, Aiken K. Solvent-free synthesis of nanoparticles. In: Török B, Dransfield T, editors. Green Chemistry. Boston, MA: Elsevier; 2017:609–646. | ||
Anjugam M, Vaseeharan B, Iswarya A, Divya M, Prabhu NM, Sankaranarayanan K. Biological synthesis of silver nanoparticles using β-1, 3 glucan binding protein and their antibacterial, antibiofilm and cytotoxic potential. Microb Pathog. 2018;115:31–40. | ||
Fariq A, Khan T, Yasmin A. Microbial synthesis of nanoparticles and their potential applications in biomedicine. J Appl Biomed. 2017;15(4):241–248. | ||
Chahardoli A, Karimi N, Fattahi A. Nigella arvensis leaf extract mediated green synthesis of silver nanoparticles: their characteristic properties and biological efficacy. Adv Powder Technol. 2018;29(1):202–210. | ||
Mittal AK, Chisti Y, Banerjee UC. Synthesis of metallic nanoparticles using plant extracts. Biotechnol Adv. 2013;31(2):346–356. | ||
Mohanraj R. Antimicrobial activities of metallic and metal oxide nanoparticles from plant extracts. In: Grumezescu AM, (editor). Antimicrobial Nanoarchitectonics. Bucharest, Romania: Elsevier; 2017:83–100. | ||
Ahmed S, Ahmad M, Swami BL, Ikram S. A review on plants extract mediated synthesis of silver nanoparticles for antimicrobial applications: a green expertise. J Adv Res. 2016;7(1):17–28. | ||
Rajeshkumar S, Bharath L. Mechanism of plant-mediated synthesis of silver nanoparticles – a review on biomolecules involved, characterisation and antibacterial activity. Chem Biol Interact. 2017;273:219–227. | ||
Arunachalam KD, Annamalai SK. Chrysopogon zizanioides aqueous extract mediated synthesis, characterization of crystalline silver and gold nanoparticles for biomedical applications. Int J Nanomedicine. 2013;8:2375. | ||
Ledezma A, Romero J, Hernández M, et al. Síntesis biomimética de nanopartículas de plata utilizando extracto acuoso de nopal (Opuntia sp.) y su electrohilado polimérico. Superf Vacío. 2014;27(4):133–140. | ||
Singh AK, Srivastava O. One-step green synthesis of gold nanoparticles using black cardamom and effect of pH on its synthesis. Nanoscale Res Lett. 2015;10(1):353. | ||
Clement BA, Goff CM, Forbes TDA. Toxic amines and alkaloids from Acacia rigidula. Phytochemistry. 1998;49(5):1377–1380. | ||
Pawar RS, Grundel E, Fardin-Kia AR, Rader JI. Determination of selected biogenic amines in Acacia rigidula plant materials and dietary supplements using LC–MS/MS methods. J Pharm Biomed Anal. 2014;88:457–466. | ||
Seigler DS. Phytochemistry of Acacia – sensu lato. Biochem Syst Ecol. 2003;31(8):845–873. | ||
Ramírez R, Lara J. Influence of native shrubs Acacia rigidula, Cercidium macrum and Acacia farnesiana on digestibility and nitrogen utilization by sheep. Small Rumin Res. 1998;28(1):39–45. | ||
Ramírez R, Ledezma-Torres R. Forage utilization from native shrubs Acacia rigidula and Acacia farnesiana by goats and sheep. Small Rumin Res. 1997;25(1):43–50. | ||
Zuo J, Mabon J. Web-based electron microscopy application software: web-EMAPS. Micro Microanal. 2004;10(S02):1000. | ||
Andrews JM. Determination of minimum inhibitory concentrations. J Antimicrob Chemother. 2001;48(suppl 1):5–16. | ||
Cavaleri J, Rankin D, Harbeck J, et al. Manual of antimicrobial susceptibility testing. Am Soc Microbiol Seattle Washington. 2005;12:53–42. | ||
Vazquez-Rodriguez A, Vasto-Anzaldo XG, Barboza Perez D, et al. Microbial competition of Rhodotorula mucilaginosa UANL-001L and E. coli increase biosynthesis of non-toxic exopolysaccharide with applications as a wide-spectrum antimicrobial. Sci Rep. 2018;8(1):798. | ||
Kugelberg E, Norström T, Petersen TK, Duvold T, Andersson DI, Hughes D. Establishment of a superficial skin infection model in mice by using Staphylococcus aureus and Streptococcus pyogenes. Antimicrob Agents Chemother. 2005;49(8):3435–3441. | ||
Rartels H, Böhmer M. Eine mikromethode 7air kreatininbestimmung. Clin Chim Acta. 1971;32(1):81–85. | ||
Fabiny DL, Ertingshausen G. Automated reaction-rate method for determination of serum creatinine with the CentrifiChem. Clin Chem. 1971;17(8):696–700. | ||
Trinder P. Determination of glucose in blood using glucose oxidase with an alternative oxygen acceptor. Ann Clin Biochem. 1969;6(1):24–27. | ||
Gornall AG, Bardawill CJ, David MM. Determination of serum proteins by means of the biuret reaction. J Biol Chem. 1949;177(2):751–766. | ||
Doumas BT, Watson WA, Biggs HG. Albumin standards and the measurement of serum albumin with bromcresol green. Clin Chim Acta. 1971;31(1):87–96. | ||
Murray R. Alanine aminotransferase. In: Kaplan LA, Pesce AJ, editors. Clinical Chemistry: Theory, Analysis, and Correlation. 2nd ed. (Chap. St Louis). St Louis, MO: The CV Mosby Company; 1989:895–898. | ||
Murray R. Aspartate aminotransferase. In: Kaplan LA, Pesce AJ, editors. Clinical Chemistry Theory, Analysis and Correlation. St Louis, MO: CV Mosby Company; 1984:1105–1108. | ||
Sadiq MB, Hanpithakpong W, Tarning J, Anal AK. Screening of phytochemicals and in vitro evaluation of antibacterial and antioxidant activities of leaves, pods and bark extracts of Acacia nilotica (L.) Del. Ind Crops Prod. 2015;77:873–882. | ||
D’Angelo JA, Zodrow EL. Chemometric study of functional groups in different layers of Trigonocarpus grandis ovules (Pennsylvanian seed fern, Canada). Org Geochem. 2011;42(9):1039–1054. | ||
Aadil KR, Barapatre A, Meena AS, Jha H. Hydrogen peroxide sensing and cytotoxicity activity of Acacia lignin stabilized silver nanoparticles. Int J Biol Macromol. 2016;82:39–47. | ||
Kumar KM, Sinha M, Mandal BK, Ghosh AR, Kumar KS, Reddy PS. Green synthesis of silver nanoparticles using Terminalia chebula extract at room temperature and their antimicrobial studies. Spectrochim Acta A Mol Biomol Spectrosc. 2012;91:228–233. | ||
Murugan K, Senthilkumar B, Senbagam D, Al-Sohaibani S. Biosynthesis of silver nanoparticles using Acacia leucophloea extract and their antibacterial activity. Int J Nanomedicine. 2014;9:2431. | ||
Viet DQ, Son Tho VD. Study on characteristics of acacia wood by FTIR and thermogrametric analysis. Viet J Chem. 2017;55(2):259. | ||
Chung I-M, Park I, Seung-Hyun K, Thiruvengadam M, Rajakumar G.Plant-mediated synthesis of silver nanoparticles: their characteristic properties and therapeutic applications. Nanoscale Res Lett. 2016;11(1):40. | ||
Banala RR, Nagati VB, Karnati PR. Green synthesis and characterization of Carica papaya leaf extract coated silver nanoparticles through X-ray diffraction, electron microscopy and evaluation of bactericidal properties. Saudi J Biol Sci. 2015;22(5):637–644. | ||
Luna C, Chávez V, Barriga-Castro ED, Núñez NO, Mendoza-Reséndez R. Biosynthesis of silver fine particles and particles decorated with nanoparticles using the extract of Illicium verum (star anise) seeds. Spectrochim Acta A Mol Biomol Spectrosc. 2015;141:43–50. | ||
Rajaram K, Aiswarya D, Sureshkumar P. Green synthesis of silver nanoparticle using Tephrosia tinctoria and its antidiabetic activity. Mater Lett. 2015;138:251–254. | ||
Perugu S, Nagati V, Bhanoori M. Green synthesis of silver nanoparticles using leaf extract of medicinally potent plant Saraca indica. Appl Nanosci. 2016;6(5):747–753. | ||
Borase HP, Salunkhe RB, Patil CD, et al. Innovative approach for urease inhibition by Ficus carica extract-fabricated silver nanoparticles: an in vitro study. Biotechnol Appl Biochem. 2015;62(6):780–784. | ||
Roopan SM, Madhumitha G, Rahuman AA, Kamaraj C, Bharathi A, Surendra T. Low-cost and eco-friendly phyto-synthesis of silver nanoparticles using Cocos nucifera coir extract and its larvicidal activity. Ind Crops Prod. 2013;43:631–635. | ||
Arunachalam KD, Annamalai SK, Hari S. One-step green synthesis and characterization of leaf extract-mediated biocompatible silver and gold nanoparticles from Memecylon umbellatum. Int J Nanomedicine. 2013;8:1307. | ||
Zhang J, Li X, Sun X, Li Y. Surface enhanced Raman scattering effects of silver colloids with different shapes. J Phys Chem B. 2005;109(25):12544–12548. | ||
Frieri M, Kumar K, Boutin A. Antibiotic resistance. J Infect Public Health. 2016;10(4):369–378. | ||
Kapil A. The challenge of antibiotic resistance: need to contemplate. Indian J Med Res. 2005;121(2):83. | ||
Singh K, Panghal M, Kadyan S, Chaudhary U, Yadav JP. Green silver nanoparticles of Phyllanthus amarus: as an antibacterial agent against multi drug resistant clinical isolates of Pseudomonas aeruginosa. J Nanobiotechnology. 2014;12(1):40. | ||
Ahmad A, Wei Y, Syed F, et al. The effects of bacteria-nanoparticles interface on the antibacterial activity of green synthesized silver nanoparticles. Microb Pathog. 2017;102:133–142. | ||
Banasiuk R, Krychowiak M, Swigon D, et al. Carnivorous plants used for green synthesis of silver nanoparticles with broad-spectrum antimicrobial activity. Arab J Chem. 2017;8:23–32. | ||
Shankar T, Karthiga P, Swarnalatha K, Rajkumar K. Green synthesis of silver nanoparticles using Capsicum frutescence and its intensified activity against E. coli. Res Effic Technol. 2017;3(3):303–308. | ||
Pattanayak S, Mollick MMR, Maity D, et al. Butea monosperma bark extract mediated green synthesis of silver nanoparticles: characterization and biomedical applications. J Saudi Chem Soc. 2015;21(6):673–684. | ||
Dakal TC, Kumar A, Majumdar RS, Yadav V. Mechanistic basis of antimicrobial actions of silver nanoparticles. Front Microbiol. 2016;7:1831. | ||
Kumar SSD, Houreld NN, Kroukamp EM, Abrahamse H. Cellular imaging and bactericidal mechanism of green-synthesized silver nanoparticles against human pathogenic bacteria. J Photochem Photobiol B. 2018;178:259–269. | ||
Salem W, Leitner DR, Zingl FG, et al. Antibacterial activity of silver and zinc nanoparticles against Vibrio cholerae and enterotoxic Escherichia coli. Int J Med Microbiol. 2015;305(1):85–95. | ||
Singla R, Soni S, Kulurkar PM, et al. In situ functionalized nanobiocomposites dressings of bamboo cellulose nanocrystals and silver nanoparticles for accelerated wound healing. Carbohydr Polym. 2017;155:152–162. | ||
Rajasekharreddy P, Rani PU, Mattapally S, Banerjee SK. Ultra-small silver nanoparticles induced ROS activated toll-pathway against Staphylococcus aureus disease in silkworm model. Mater Sci Eng C. 2017;77:990–1002. | ||
Anjum S, Abbasi BH. Thidiazuron-enhanced biosynthesis and antimicrobial efficacy of silver nanoparticles via improving phytochemical reducing potential in callus culture of Linum usitatissimum L. Int J Nanomedicine. 2016;11:715. | ||
Hussain M, Raja NI, Iqbal M, Aslam S. Applications of plant flavonoids in the green synthesis of colloidal silver nanoparticles and impacts on human health. Iran J Sci Technol Trans A Sci. 2017;2:1–12. | ||
Murray R, Steed P, Elson H. The location of the mucopeptide in sections of the cell wall of Escherichia coli and other Gram-negative bacteria. Can J Microbiol. 1965;11(3):547–560. | ||
Shockman GD, Barren J. Structure, function, and assembly of cell walls of Gram-positive bacteria. Annu Rev Microbiol. 1983;37(1):501–527. | ||
Dibrov P, Dzioba J, Gosink KK, Häse CC. Chemiosmotic mechanism of antimicrobial activity of Ag+ in Vibrio cholerae. Antimicrob Agents Chemother. 2002;46(8):2668–2670. | ||
Pal S, Tak YK, Song JM. Does the antibacterial activity of silver nanoparticles depend on the shape of the nanoparticle? A study of the Gram-negative bacterium Escherichia coli. Appl Environ Microbiol. 2007;73(6):1712–1720. | ||
Holt KB, Bard AJ. Interaction of silver (I) ions with the respiratory chain of Escherichia coli: an electrochemical and scanning electrochemical microscopy study of the antimicrobial mechanism of micromolar Ag+. Biochemistry. 2005;44(39):13214–13223. | ||
Kaviya S, Santhanalakshmi J, Viswanathan B. Green synthesis of silver nanoparticles using Polyalthia longifolia leaf extract along with D-sorbitol: study of antibacterial activity. J Nanotechnol. 2011;2011:5. | ||
Das B, Dash SK, Mandal D, et al. Green synthesized silver nanoparticles destroy multidrug resistant bacteria via reactive oxygen species mediated membrane damage. Arab J Chem. 2017;10(6):862–876. | ||
Kim JS, Kuk E, Yu KN, et al. Antimicrobial effects of silver nanoparticles. Nanomedicine. 2007;3(1):95–101. | ||
Ivask A, ElBadawy A, Kaweeteerawat C, et al. Toxicity mechanisms in Escherichia coli vary for silver nanoparticles and differ from ionic silver. ACS Nano. 2013;8(1):374–386. | ||
Klaassen CD, Amdur MO. Casarett and Doull’s Toxicology: The Basic Science of Poisons. New York: McGraw-Hill, Health Professions Division, USA 1996. | ||
Vasquez RD, Apostol JG, de Leon JD, et al. Polysaccharide-mediated green synthesis of silver nanoparticles from Sargassum siliquosum JG Agardh: assessment of toxicity and hepatoprotective activity. OpenNano. 2016;1:16–24. | ||
Roy N, Gaur A, Jain A, Bhattacharya S, Rani V. Green synthesis of silver nanoparticles: an approach to overcome toxicity. Environ Toxicol Pharmacol. 2013;36(3):807–812. | ||
Zhang XF, Liu ZG, Shen W, Gurunathan S. Silver nanoparticles: synthesis, characterization, properties, applications, and therapeutic approaches. Int J Mol Sci. 2016;17(9):pii:E1534. | ||
Chidurala SC, Kalagadda VR, Tambur P. Antimicrobial activity of pure Cu nano particles synthesized by surfactant varied chemical reduction method. Environmental Nanotechnology, Monitoring & Management. 2016;6:88–94. | ||
Bindhu M, Umadevi M. Antibacterial activities of green synthesized gold nanoparticles. Materials Letters. 2014;120:122–125. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.