Back to Journals » Breast Cancer: Targets and Therapy » Volume 15
Hsa_circ_0007823 Overexpression Suppresses the Progression of Triple-Negative Breast Cancer via Regulating miR-182-5p-FOXO1 Axis
Authors Yu J, Wang H, Shen W, Zhou Y, Cui J, Li H, Gao B
Received 17 April 2023
Accepted for publication 10 October 2023
Published 18 October 2023 Volume 2023:15 Pages 695—708
DOI https://doi.org/10.2147/BCTT.S417547
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Pranela Rameshwar
Jinling Yu,1 Haofeng Wang,1 Weida Shen,1 Yingzi Zhou,2 Jing Cui,1 Haichuan Li,3 Beimin Gao1
1Department of Breast Surgery, Shanghai Changning Maternity and Infant Health Hospital, East China Normal University, Shanghai, 200050, People’s Republic of China; 2Department of Pathology, Shanghai Changning Maternity and Infant Health Hospital, East China Normal University, Shanghai, 200050, People’s Republic of China; 3Department of Laboratory, Shanghai Changning Maternity and Infant Health Hospital, East China Normal University, Shanghai, 200050, People’s Republic of China
Correspondence: Beimin Gao; Haichuan Li, Shanghai Changning Maternity and Infant Health Hospital, East China Normal University, No. 786, Yuyuan Road, Changning District, Shanghai, 200050, People’s Republic of China, Tel +86-21-62288686-2401, Email [email protected]; [email protected]
Background: This study aimed to analyze the specific expression of hsa_circ_0007823 in triple-negative breast cancer (TNBC) and explore the roles and related molecular mechanisms of hsa_circ_0007823 in TNBC.
Materials and Methods: Relative hsa_circ_0007823 levels in TNBC tissues and cell lines were examined by reverse transcription-quantitative polymerase chain reaction. The value of hsa_circ_0007823 levels was evaluated in patients’ clinicopathological characteristics and prognostic prediction. A dual-luciferase reporter assay was used to determine the relationship between hsa_circ_0007823, miR-182-5p, and FOXO1. The effect of circ_0007823 overexpression on the growth of TNBC cells was investigated in vitro and in vivo.
Results: Lower levels of hsa_circ_0007823 were found in TNBC tissues and cell lines and were closely associated with lymph node metastasis, poorer overall and disease-free survival rates. MiR-182-5p was significantly up-regulated, whereas FOXO1 was down-regulated in TNBC cell lines. The miR-182-5p inhibition up-regulated FOXO1 in TNBC cells. Dual-luciferase reporter assays showed that hsa_circ_0007823, miR-182-5p, and FOXO1 interacted with each other. Overexpression of circ_0007823 significantly inhibited the viability, migration, and invasion of TNBC cell lines, but promoted apoptosis. In vivo experiments showed that circ_0007823 overexpression inhibited tumor growth and down-regulated miR-182-5p and up-regulated FOXO1.
Conclusion: Hsa_circ_0007823 overexpression could suppress the growth, invasion, and migration of TNBC cells, and inhibit tumor growth by regulating miR-182-5p/FOXO1.
Keywords: triple-negative breast cancer, hsa_circ_0007823, miR-182, FOXO1, biological functional analysis
Introduction
Breast cancer is the most common cancer, with more than 2.26 million new cases and approximately 685,000 deaths worldwide.1 Breast cancer is classified into luminal, HER2-amplified, and triple-negative breast cancer (TNBC).2 Luminal breast cancer, the most common subtype, has the best prognosis and benefits from targeted endocrine therapy due to hormone receptor expression as well as HER2-amplified breast cancer benefits from anti-HER2 therapy.3 However, TNBC is a biologically and clinically heterogeneous disease that differs from other subtypes, TNBC develops metastases earlier after diagnosis, with higher brain, liver, and lung metastasis rates, and relatively poor outcomes and prognosis.4 Endocrine therapy, molecular-targeted therapy, and chemotherapy, which are effective for other subtypes, are not effective for treating TNBC, and standardized treatment protocols for TNBC are lacking.5 Three randomized clinical trials of combination treatment strategies using immunomodulators (monoclonal antibodies) and chemotherapeutic agents are underway.6,7 Therefore, developing new therapeutic regimens and targets for TNBC is urgently required.
Advances in sequencing technologies have enabled the identification of a large class of biomolecules called non-coding RNAs. Circular RNAs (circRNAs), a class of endogenous non-coding RNAs that are covalent, single-stranded, closed-loop structures that are not easily degraded by RNA enzymes and nucleic acid exonucleases, and are highly stable.8,9 CircRNAs are widely and abundantly expressed in various mammalian tissues and cells.10 In addition, circRNAs exhibit high evolutionary conservation and tissue, cellular, and developmental stage specificity.11,12 CircRNAs can also be released into various body fluids via exosomes; they are stably present, circulate in body fluids, and are recognized by recipient cells.13 Their reported mechanisms of action of circRNAs include acting as microRNA (miRNA) sponges, translation templates for proteins/peptides, and regulating the expression of parental genes.14 Dysregulated expression of circRNAs inevitably leads to the development of various diseases, including cancer.15,16 Liu et al17 showed that circEZH2 was upregulated in the liver metastases of breast cancer and could predict worse outcomes in patients with breast cancer. Another study demonstrated that circKIF4A was overexpressed in breast cancer cell lines and tissues, and the inhibition of its expression could suppress breast cancer growth and metastasis.18 These reports confirmed the presence of specific circRNA sets in different breast cancer subtypes. A previous study used RNA sequencing technology to identify 26 up-regulated and 113 down-regulated circRNAs in TNBC, which were closely associated with the brain-derived neurotrophic factor receptor signaling pathway, as well as AMPK and TGF-β signaling pathways.19 In addition, increasing dysregulated circRNAs in TNBC have been identified as potential tumor markers or therapeutic targets.20 However, the specific functions and mechanisms of action of circRNAs in TNBC remain unclear.
Our previous studies confirmed that miR-182-5p levels were higher in TNBC tissues and could promote cell proliferation and migration in TNBC by down-regulating forkhead box F2 (FOXF2).21,22 Thereafter, we performed circRNA sequencing on the samples of TNBC and adjacent tissues to establish the expression profiles of circRNAs of TNBC and found five circRNAs (chr6:131973682-132047340+, hsa_circ_0005982, hsa_circ_0007823 (circUSP42), hsa_circ_0002032, and hsa_circ_0001777) that served as sponges for miR-182-5p. After further validation, hsa_circ_0007823 (circUSP42) was found to be significantly down-regulated in the TNBC, and bound strongly with miR-182-5p.23 Furthermore, the forkhead box O (FOXO) transcription factor family is frequently mutated, deleted, or amplified in various human cancers, making it an attractive candidate for treatment.24 It has been reported that miR-182-5p could inhibit the growth of T cells by inhibiting FOXO1 expression in breast cancer, indicating that miR-182-5p may interact with FOXO1,25 but their relationship needs to be further investigated. Therefore, we explored in vivo and in vitro, the roles and underlying mechanisms of the hsa_circ_0007823-miR-182-5p-FOXO1 axis in TNBC occurrence and development. The study results contribute to understanding the possible molecular mechanisms of TNBC pathogenesis and providing potential targets and pathways for TNBC treatment.
Materials and Methods
Samples
Tumors and adjacent normal tissues from 30 patients with TNBC who underwent modified radical surgery at the Shanghai Changning Maternity and Infant Health Care Hospital were collected and immediately preserved in RNAlater (Ambion). The inclusion criteria were as follows: (1) female patients aged 31–73 years old with a definite diagnosis of TNBC; (2) patients who underwent modified radical surgery for breast cancer; (3) patients with complete clinicopathological data; and (4) patients who provided informed consent. The exclusion criteria were as follows: (1) patients with preoperative radiotherapy or neoadjuvant chemotherapy; (2) patients with distant metastasis; and (3) patients with special pathological types, such as invasive lobular, medullary, mucinous, invasive micropapillary, and metaplastic carcinomas. The study complied with the Declaration of Helsinki and was approved by the Ethics Committee of Shanghai Changning Maternity and Infant Health Care Hospital (approval no. CNFBLLKT-2020002). Written informed consent was obtained from all the participants.
Cell Culture and Transfection
The TNBC cell lines MDA-MB-231, MDA-MB-468, BT-549, human control mammary epithelial cells MCF10A, and 293T cells were purchased from the Cell Bank of the Chinese Academy of Science (Shanghai, China). The MDA-MB-231, MDA-MB-468, and 293T cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Gibco, Invitrogen, USA) containing 10% fetal bovine serum (FBS, Gibco, Invitrogen, USA), penicillin (100 U/mL, Gibco, Invitrogen, USA), and streptomycin (100 μg/mL, Gibco, Invitrogen, USA). The BT-549 cells were cultured in RPMI-1640 medium with 10% FBS, and 1% penicillin/streptomycin. The MCF10A cells were cultured in DMEM/F12 medium supplemented with 10% FBS and 1% penicillin/streptomycin. All the cells were maintained in an incubator with 5% CO2 at 37 °C.
The hsa_circ_0007823 sequence was synthesized using General Biosystems (Chuzhou, Anhui, China) and cloned into pLCDH plasmids (Yanzai Biotechnology, Shanghai, China) to obtain the pLCDH-hsa_circ_0007823 plasmids. The miR-182-5p inhibitor and negative control (NC) inhibitor were purchased from Ribobio (Guangzhou, China). Cell transfection was performed using Lipofectamine 3000 (Thermo Fisher Scientific, Carlsbad, CA, USA) according to the manufacturer’s instructions. After culturing for 24 h, the cell transfection efficiency was assessed by determining the levels of circ_0007823 and miR-182-5p using quantitative real-time polymerase-chain reaction (qRT-PCR).
qRT-PCR
Total RNA was extracted from the different cell lines and cells with different treatments or tissue samples using RNAiso Plus (TAKARA, Dalian, China) according to the standard methods. Then, cDNA was synthesized using PrimeScript™ RT Master Mix Kit (TAKARA) for examining miRNA levels, or by PrimeScript™ II 1st Strand cDNA Synthesis Kit (TAKARA) for examining mRNA or circRNA expressions. Amplification was performed using the ViiA 7 Real-time PCR System (Thermo Fisher Scientific). The primer sequences used are listed in Table 1. The expression of circRNA and mRNA was quantified using GAPDH as an internal reference gene, and miRNA expression was normalized to U6 expression using the 2−ΔΔCt method.
 |
Table 1 The Primer Sequences |
Cell Viability and Apoptosis Assay
Cell viability was determined using the cell Counting Kit-8 (CCK-8). Cells with different treatments (approximately 1 × 104) were cultured for 48 h, and 10 µL of CCK-8 reagent (Dojindo, Japan) was added, followed by incubation for another 2 h. The optical density (OD) was measured at 450 nm using a microplate reader.
Following the manufacturer’s protocols, cell apoptosis was examined using an Annexin V-FITC apoptosis detection kit (BD Pharmingen, CA, USA). Briefly, the cells subjected to different treatments were harvested and centrifuged at 1000 rpm for 5 min. The cells were resuspended with Annexin V-FITC (195 μL), and 5 μL Annexin V-FITC and 5 μL PI (50 μg/mL) were added. After incubation in the dark at 25 °C for 20 min, the images were acquired using a BD FACSCalibur flow cytometer (BD Pharmingen, CA, USA), and cell apoptosis rates were analyzed.
Transwell and Scratch Assays
Transwell chambers (8-µm pore size; Guangzhou Jet Bio-Filtration Co. Ltd., Guangzhou, China) pre-coated with or without a layer of Matrigel (extra cellular matrix gel; Corning, USA) were used to determine cell migration and invasion. Briefly, approximately 4 × 104 cells suspended in 200 μL of DMEM supplemented with 0.1% FBS were transferred into the upper chamber of Transwell plates, with the lower chamber containing the complete medium. After incubation for 48 h, all media were removed. Next, 4% paraformaldehyde was added to the chambers and incubated for 10 min, followed by incubation with 0.1% crystal violet for 20 min. After removing excess dye and air-drying, the images were observed under an inverted microscope (IX70, Olympus Corporation, Japan), and the relative numbers of the migrating and invading cells were counted.
In addition, a scratch test was performed to determine cell migration. Briefly, cells subjected to different treatments were used for the scratch test as follows: a sharp-pointed tool was used to scratch the horizontal line behind the cell culture medium, then the cell culture medium was removed. After washing twice with PBS, serum-free medium was added and cultured for 48 h, and the cells were photographed at 0 h and 48h, respectively.
Dual-Luciferase Reporter Assay
The hsa_circ_0007823 sequence was cloned into the polyclonal site downstream of the translation termination codon of Renilla luciferase (Rluc) in the psiCHECK2 plasmid to obtain the psiCHECK2-hsa_circ_0007823 plasmid. The interaction of hsa_circ_0007823 with the interfering RNA present in the system affected Rluc expression. PsiCHECK2-hsa_circ_0007823, psiCHECK2, miR-NC, or miR-182-5p mimics were co-transfected into 293T cells. After incubation for 48 h, relative Rluc activity was measured using a dual-luciferase reporter system (TECAN, Männedorf, Switzerland).
PGL3 vectors (Promega, Wisconsin, USA) were inserted with wild-type FOXO1 mRNA 3’ UTR sequence or FOXO1 mRNA 3’ UTR sequence with a putative binding sequence mutation. The pGL3 recombinant plasmid, miR-NC, and the miR-182-5p mimics were co-transfected into MDA-MB-231 cells. After incubation for 48 h, firefly luciferase activity was inspected using a microplate reader (Perkin-Elmer) and normalized to Rluc activity.
Tumor Formation in Nude Mice
MDA-MB-231 cells and MDA-MB-231 cells overexpressing circ_0007823 were harvested and centrifuged at 1000 rpm for 5 min. After washing twice with PBS, the cells were resuspended in a serum-free medium, and the concentration of the cells was adjusted to 5 × 107 cells/mL for use.
Ten SPF BALB/c female nude mice aged 4–6 weeks were purchased from Shanghai SLAC Laboratory Animal Co., Ltd. (Shanghai, China). All the mice were maintained under controlled temperature (24 ± 2°C) and humidity (50 ± 5%), with a 12 h light/dark cycle. After seven days of acclimatization, the mice were randomly divided into two groups (n = 5): control and OE-circ_0007823 groups. The mice in the control and OE-circ_0007823 groups were subcutaneously injected with 100 μL MDA-MB-231 cells and 100 μL MDA-MB-231 cells with circ_0007823 overexpression H1975 cells in the right forearm underarm, respectively. During the experiment, the mice had ad libitum access to food and water. After tumor inoculation, the subcutaneous graft volume in nude mice was measured weekly. After four weeks, all mice were sacrificed by cervical dislocation, and the tumors were stripped, weighed, and photographed. Subsequently, the tumor tissues from each group were subjected to qRT-PCR and Western blot. All the animal experiments were performed in accordance with the National Medical Advisory Committee (NMAC) guidelines, using procedures approved by the Institutional Animal Care and Use Committee at Shanghai Changning Maternity and Infant Health Care Hospital.
Western Blot
Proteins were extracted, separated using sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and transferred onto polyvinylidene difluoride (PVDF) membranes. The membranes were blocked and incubated with primary antibodies against FOXO1 (18592-1-AP, Proteintech) and GAPDH (10494-1-AP; Proteintech) at 4 °C overnight and then incubated with the secondary antibody (111-035-003, Jackson ImmunoResearch) at 37 °C for 2 h. ECL chemiluminescence reagent (Beyotime, Shanghai, China) was used to visualize the bands.
Statistical Analysis
Data were analyzed using SPSS 22.0 and are presented as mean ± standard deviation. Comparisons between groups were performed using t-tests or one-way analysis of variance. Pearson’s correlation test was used to analyze pairwise expression correlations. Kaplan–Meier plots and Log rank tests were used to determine the correlation between hsa_circ_0007823 levels, disease-free survival (DFS), and overall survival (OS). P < 0.05 indicates statistical significance.
Results
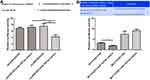
Low Expression of Hsa_circ_0007823 Was Associated with Metastasis and Prognosis of TNBC
The expression levels of hsa _circ_0007823 were determined by qRT-PCR. It was found that hsa _circ_0007823 levels were down-regulated in the TNBC tissues (5.97 ± 3.07) compared with the adjacent tissues (20.32 ± 5.42) (t = 16.55, P < 0.001, Figure 1A). After analyzing the relationships between hsa_circ_0007823 and TNBC clinical indicators, the hsa_circ_0007823 level was significantly correlated with lymph node metastasis (P = 0.014) but not with age, menopausal status, histological grade, TNM staging, and Ki-67 expression (P > 0.05, Table 2). The hsa_circ_0007823 level was examined in the lymph node metastasis tissues, and it was found that the hsa _circ_0007823 levels were significantly lower in the lymph node metastasis tissues than in the non-metastatic tissues (P < 0.01, Figure 1B). Furthermore, patients with low hsa_circ_0007823 levels had significantly poorer OS (P = 0.0003, Figure 1C) and DFS (P = 0.0002, Figure 1D) in comparison with the patients with high hsa_circ_0007823 levels. These results implied that low levels of hsa_circ_0007823 may be closely associated with poor prognosis and lymph node metastasis in TNBC.
 |
Table 2 Correlation Between Circ_0007823 Levels and TNBC Clinical Indicators |
Hsa_circ_0007823, miR-182-5p, and FOXO1 Interacted with Each Other
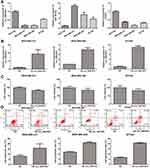
To elucidate the relationships of hsa_circ_0007823, miR-182-5p, and FOXO1, miRanda was used to predict the binding site of hsa_circ_0007823 and miR-182-5p, as well as TargetScan was employed to predict that bases 250–271 of the FOXO1 mRNA 3’ UTR may be the potential target regions for miR-182-5p. These relationships were confirmed using a dual luciferase reporter assay. There were no significant differences in the luciferase activity among the cells co-transfected with empty plasmids and miR-NC, the cells co-transfected with empty plasmids and miR-182-5p mimics, and the cells co-transfected with psiCHECK2-hsa_circ_0007823 and miR-NC (P > 0.05); whereas the luciferase activity in the cells co-transfected with psiCHECK2-hsa_circ_0007823 and miR-182-5p mimics was significantly decreased compared with the cells co-transfected with empty plasmids and miR-182-5p mimics, or the cells co-transfected with psiCHECK2-hsa_circ_0007823 and miR-NC (P < 0.05, Figure 2A). These results indicated that hsa_circ_0007823 could interact with miR-182-5p.
Furthermore, in the mutated FOXO1 plasmids, the luciferase activity was not significantly changed by miR-182 mimics compared with the NC (P > 0.05). In contrast, in the wild-type FOXO1 plasmids, the luciferase activity after co-transfection with miR-182-5p mimics was evidently reduced relatively to the NC (P < 0.05, Figure 2B). These results indicated that FOXO1 was the target of miR-182-5p. All the results suggested that circ_0007823, miR-182-5p, and FOXO1 could interact with each other.
Effects of Hsa_circ_0007823 on the Viability and Apoptosis of TNBC Cell Lines
The expression levels of circ_0007823, miR-182-5p, and FOXO1 were determined in TNBC cell lines (MDA-MB-231, MDA-MB-468, and BT549). Compared to the control cells (MCF10A), the levels of circ_0007823 and FOXO1 were significantly decreased in the TNBC cell lines (MDA-MB-231, MDA-MB-468, and BT549). In contrast, the miR-182-5p level was significantly increased in the TNBC cell lines (P < 0.05, Figure 3A). To investigate the roles of circ_0007823 in TNBC, TNBC cell lines overexpressing circ_0007823 were constructed, and cell growth was examined. It was found that after transfected with OE-circ_0007823 plasmids, the circ_0007823 levels in the MDA-MB-231, MDA-MB-468, and BT549 cells were markedly increased (P < 0.05, Figure 3B), which indicated that the MDA-MB-231, MDA-MB-468, and BT549 cells overexpressing circ_0007823 were successfully established.
The viability and apoptosis of the TNBC cell lines were examined after transfection with the OE-circ_0007823 plasmids. Relative to the NC cells, that is, MDA-MB-231, MDA-MB-468, and BT549 cells, transfected with OE-circ_0007823 plasmids significantly inhibited the viability of the corresponding MDA-MB-231, MDA-MB-468, and BT549 cells (P < 0.05, Figure 3C). Additionally, the results of flow cytometry showed that the total cell apoptosis rates in the TNBC cells with circ_0007823 overexpression were evidently higher than that in their corresponding normal TNBC cells (P < 0.05, Figure 3D). All these suggested that circ_0007823 was down-regulated in the TNBC cells, as well as that its overexpression could inhibit the viability of TNBC cells, whereas promote their apoptosis.
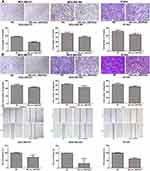
Effects of Hsa_circ_0007823 on the Invasion and Migration of TNBC Cell Lines
The effects of circ_0007823 on the migration and invasion of TNBC cell lines were also tested. It was found that the cell number of invasion in the TNBC cell lines transfected with OE-circ_0007823 plasmids was significantly lower than that in the control TNBC cell lines (P < 0.05, Figure 4A). Furthermore, both Transwell and scratch tests were used to determine cell migration. Transwell assay showed that OE-circ_0007823 plasmids observably reduced the cell number of migration relative to the control cells (P < 0.05, Figure 4B). Scratch test showed that compared to the control cell lines, the wound healing of the cell lines with circ_0007823 overexpression was signally lessened (P < 0.05, Figure 4C). Taken together, circ_0007823 overexpression could suppress the invasion and migration of the MDA-MB-231, MDA-MB-468, and BT549 cells.
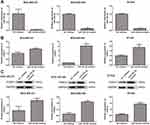
Expression of FOXO1 in the TNBC Cell Lines Transfected with miR-182-5p Inhibitor
Further, we constructed the TNBC cell lines with miR-182-5p silencing using miR-182-5p inhibitor and found that after transfection, the levels of miR-182-5p in the TNBC cell lines were significantly decreased compared with the cells with NC inhibitor (P < 0.01, Figure 5A), which implied that the TNBC cell lines with miR-182-5p knockdown were successfully constructed and could be used for subsequent experiments. Then, the expression of FOXO1 was determined using qRT-PCR and Western blot. It was obvious that compared with the NC inhibitor-transfected cells, FOXO1 mRNA expression and protein expression were both significantly up-regulated after transfected with miR-182-5p inhibitor (P < 0.01, Figure 5B and C).
Overexpression of Hsa_circ_0007823 Inhibited the Growth of TNBC Tumor
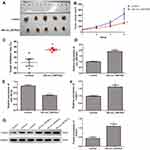
Finally, the roles of circ_0007823 in TNBC were confirmed using in vivo experiments. MDA-MB-231 cells and MDA-MB-231 cells overexpressing circ_0007823 were, respectively, injected to nude mice, and the tumor of each mouse was observed. After four-weeks of feeding, the tumors in the mice injected with circ_0007823-overexpressed MDA-MB-231 cells were smaller than those in the mice injected with MDA-MB-231 cells (Figure 6A). With the increasing of feeding time, the tumor volume in each group was gradually increased, while the tumor size in the OE-circ_0007823 group was significantly lower than that in the control group (P < 0.06, Figure 6B). Additionally, the tumor inhibition rate in the OE-circ_0007823 group was memorably higher than that in the control group (P < 0.06, Figure 6C). These hinted that circ_0007823 overexpression could inhibit the growth of TNBC tumor.
The levels of circ_0007823, miR-182-5p, and FOXO1 were determined in the tumor samples of each group. Compared with the control mice, the expression of circ_0007823 in the OE-circ_0007823 group was significantly up-regulated (P < 0.01, Figure 6D); whereas the level of miR-182-5p was evidently decreased (P < 0.01, Figure 6E). For FOXO1, its mRNA expression and protein expression were both significantly up-regulated in the OE-circ_0007823 tissue samples (P < 0.05, Figure 6F and G).
Discussion
In most countries, female breast cancer ranks first in morbidity and mortality among all cancer types, and TNBC is a subtype of breast cancer that is difficult to cure.26 It has been reported that dysregulation of non-coding RNAs, including circRNAs, miRNAs, and related key proteins, drives breast cancer development and promotes cancer progression.27 Exploration of these key dysregulated factors is of great importance. These factors may serve as early biomarkers for the early diagnosis of TNBC, which will directly impact patient treatment and survival. Previous studies found that hsa_circ_0006743, hsa_circ_0002496, hsa_circ_0005046, and hsa_circ_0001791 have been identified to be abnormally expressed in early breast cancer, thereby may be valuable for the early diagnosis of breast cancer.28,29 These factors may be also used to identify the biological features of tumors and help to better manage cancers. Wang et al reported that the high level of circ-UBE2D2 was found to be closely associated with aggressive clinical features of breast cancer.30 In addition, stratifying patients with breast cancer according to these factors provides a more appropriate therapeutic strategy or personalized treatment for patients, as well as targeting these factors may be useful for the treatment of TNBC.31 Breast cancer, including TNBC, can be treated using techniques such as antisense oligonucleotides or CRISPR/Cas to downregulate or suppress circRNAs upregulated in breast cancer and exert pro-cancer effects.32,33 Some circRNAs are downregulated in breast cancer tissues, and function as tumor suppressors.34,35 Nanoparticles containing specific circRNAs have been shown to inhibit tumor growth and metastasis.36,37 Finally, the identification of the expression levels of these factors may be used to analyze tumor response to therapy and predict further cancer progression (growth and metastasis) and prognosis. Therefore, continued identification of new validated biomarkers is of significant clinical value.
In this study, hsa_circ_0007823 was significantly down-regulated in the TNBC tissues and TNBC cell lines, which were in accordance with our previous sequencing data of TNBC tissues.23 Lower levels of hsa_circ_0007823 were also closely associated with lymph node metastasis, and poor overall survival and disease-free survival of TNBC. Furthermore, in vitro and in vivo experiments found that hsa_circ_0007823 overexpression significantly inhibited viability, migration and invasion of TNBC cell lines, whereas enhanced their apoptosis as well as suppressed the tumor growth. Chen et al38 demonstrated that circ_0000285 level was higher in the cervical cancer tissues, as well as knockdown of circ_0000285 could suppress the growth and migration of cervical cancer cells and inhibit the formation and transfer of cervical cancer in nude mice through regulating FUS. Another research manifested that circRNA_102231 was up-regulated in the gastric cancer tissues and plasma, as well as circRNA_102231 knockdown could inhibit gastric cancer cell proliferation and invasion both in vitro and in vivo; which implied that circRNA_102231 may be used as a potential biomarker and therapeutic target for gastric cancer.39 Taken together, we speculate that hsa_circ_0007823 may be a predictive and prognostic biomarker for TNBC, and overexpression of circ_0007823 may be beneficial for TNBC treatment. However, large retrospective or prospective clinical cohort studies with expanded inclusion sample sizes are necessary, together with an analysis of the differential expression of hsa_circ_0007823 in early stage TNBC biological characteristics. In addition, the analysis of the specific levels of hsa_circ_0007823 in blood samples from patients with TNBC may be promising.
Further, in order to understand the molecular mechanisms of circ_0007823 in TNBC, the target miRNA and mRNA were predicted, and dual-luciferase reporter gene assay showed that circ_0007823, miR-182-5p, and FOXO1 could interact with each other. RT-qPCR and Western blot showed that down-regulation of miR-182-5p could increase the FOXO1 level, as well as circ_0007823 overexpression could repress the miR-182-5p level, whereas up-regulate FOXO1. miR-182-5p was found to be highly expressed in the tumor tissues and plasma of patients with breast cancer and was considered to have an early breast cancer diagnostic value.40 Increasing evidence showed that miR-182-5p could promote growth, metastasis, angiogenesis, and tamoxifen and veliparib resistance in breast cancer.41,42 In addition, down-regulation of FOXO1 expression and promoter hypermethylation has been identified in breast cancer tissues, and correlated with tumor size and lymph node metastasis.43 Patients with breast cancer with high FOXO1 expression have better prognosis.44 A study of Zeng et al reported that FOXO1 was associated with immune cell infiltration, immune checkpoint expression, and response to immunotherapy in breast cancer.45 It has been found that FOXO1 expression was also responsible for stemness and sensitivity of breast cancer cells to chemotherapeutic agents, such as tamoxifen and paclitaxel.46,47 Combined with our results, it can be inferred that hsa_circ_0007823 overexpression may inhibit the progression of TNBC through regulating the miR-182-5p/FOXO1 axis. However, in addition to miR-182-5p and FOXO1, circ_0007823 may also adsorb other miRNAs or be involved in TNBC progression through other molecular mechanisms, which requires further research.
Conclusions
Lower levels of hsa_circ_0007823 were found in TNBC tissues and closely associated with metastasis and poor outcomes. Additionally, overexpression of circ_0007823 may suppress the growth, invasion, and migration of TNBC cells, and inhibit tumor growth in nude mice via regulating miR-182-5p/FOXO1, thereby hindering the development and metastasis of TNBC. These findings indicate that circ_0007823 may be a predictive and prognostic biomarker for TNBC, as well as provide a basis for circ_0007823/miR-182-5p/FOXO1 as novel therapeutic targets and pathways for TNBC management.
Abbreviations
qRT-PCR, quantitative real time-PCR, TNBC, triple-negative breast cancer; circRNA, circular RNA; FOXO, forkhead box O; NC, negative control; CCK-8, cell counting kit-8; Rluc, renilla luciferase; OS, overall survival; DFS, disease-free survival; miRNA, microRNA.
Funding
This study was supported by Health Industry Clinical Research Project of Shanghai Municipal Health Commission (No.202040144).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer Statistics, 2021. CA Cancer J Clin. 2021;71(1):7–33. doi:10.3322/caac.21654
2. Mavrommati I, Johnson F, Echeverria GV, Natrajan R. Subclonal heterogeneity and evolution in breast cancer. NPJ Breast Cancer. 2021;7(1):155. doi:10.1038/s41523-021-00363-0
3. Chu J, Li Y, He M, et al. Zinc finger and SCAN domain containing 1, ZSCAN1, is a novel stemness-related tumor suppressor and transcriptional repressor in breast cancer targeting TAZ. Front Oncol. 2023;13:1041688. doi:10.3389/fonc.2023.1041688
4. Xiao W, Zheng S, Yang A, et al. Breast cancer subtypes and the risk of distant metastasis at initial diagnosis: a population-based study. Cancer Manag Res. 2018;10:5329–5338. doi:10.2147/CMAR.S176763
5. Derakhshan F, Reis-Filho JS. Pathogenesis of triple-negative breast cancer. Annu Rev Pathol. 2022;17(1):181–204. doi:10.1146/annurev-pathol-042420-093238
6. Schmid P, Rugo HS, Adams S, et al. Atezolizumab plus nab-paclitaxel as first-line treatment for unresectable, locally advanced or metastatic triple-negative breast cancer (IMpassion130): updated efficacy results from a randomised, double-blind, placebo-controlled, Phase 3 trial. Lancet Oncol. 2020;21(1):44–59. doi:10.1016/S1470-2045(19)30689-8
7. Cortes J, Cescon DW, Rugo HS, et al. Pembrolizumab plus chemotherapy versus placebo plus chemotherapy for previously untreated locally recurrent inoperable or metastatic triple-negative breast cancer (KEYNOTE-355): a randomised, placebo-controlled, double-blind, phase 3 clinical trial. Lancet. 2020;396(10265):1817–1828. doi:10.1016/S0140-6736(20)32531-9
8. Vo JN, Cieslik M, Zhang Y, et al. The landscape of circular RNA in cancer. Cell. 2019;176(4):869–881 e13. doi:10.1016/j.cell.2018.12.021
9. Patop IL, Wust S, Kadener S. Past, present, and future of circRNAs. EMBO J. 2019;38(16):e100836. doi:10.15252/embj.2018100836
10. Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157. doi:10.1261/rna.035667.112
11. Wang PL, Bao Y, Yee MC, et al. Circular RNA is expressed across the eukaryotic tree of life. PLoS One. 2014;9(6):e90859. doi:10.1371/journal.pone.0090859
12. Rybak-Wolf A, Stottmeister C, Glazar P, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2015;58(5):870–885. doi:10.1016/j.molcel.2015.03.027
13. Wang Y, Liu J, Ma J, et al. Exosomal circRNAs: biogenesis, effect and application in human diseases. Mol Cancer. 2019;18(1):116. doi:10.1186/s12943-019-1041-z
14. Zang J, Lu D, Xu A. The interaction of circRNAs and RNA binding proteins: an important part of circRNA maintenance and function. J Neurosci Res. 2020;98(1):87–97. doi:10.1002/jnr.24356
15. Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32(5):453. doi:10.1038/nbt.2890
16. Chen LL. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat Rev Mol Cell Biol. 2020;21(8):475–490. doi:10.1038/s41580-020-0243-y
17. Liu P, Wang Z, Ou X, et al. The FUS/circEZH2/KLF5/ feedback loop contributes to CXCR4-induced liver metastasis of breast cancer by enhancing epithelial-mesenchymal transition. Mol Cancer. 2022;21(1):198. doi:10.1186/s12943-022-01653-2
18. Huang J, Deng X, Chen X, et al. Circular RNA KIF4A promotes liver metastasis of breast cancer by reprogramming glucose metabolism. J Oncol. 2022;2022:8035083. doi:10.1155/2022/8035083
19. Li X, Zhang S, Sun S, et al. Prediction and screening of circRNA in triple-negative breast cancer. Am J Transl Res. 2022;14(11):8049–8063.
20. Lyu L, Zhang S, Deng Y, et al. Regulatory mechanisms, functions, and clinical significance of CircRNAs in triple-negative breast cancer. J Hematol Oncol. 2021;14(1):41. doi:10.1186/s13045-021-01052-y
21. Yu J, Shen W, Gao B, Zhao H, Xu J, Gong B. MicroRNA-182 targets FOXF2 to promote the development of triple-negative breast cancer. Neoplasma. 2017;64(2):209–215. doi:10.4149/neo_2017_206
22. Zhang X, Ma G, Liu J, Zhang Y. MicroRNA-182 promotes proliferation and metastasis by targeting FOXF2 in triple-negative breast cancer. Oncol Lett. 2017;14(4):4805–4811. doi:10.3892/ol.2017.6778
23. Yu J, Shen W, Xu J, Gong B, Gao B, Zhu J. circUSP42 is downregulated in triple-negative breast cancer and associated with poor prognosis. Technol Cancer Res Treat. 2020;19:1533033820950827. doi:10.1177/1533033820950827
24. Xie J, Zhang J, Tian W, et al. The pan-cancer multi-omics landscape of FOXO family relevant to clinical outcome and drug resistance. Int J Mol Sci. 2022;23(24):15647. doi:10.3390/ijms232415647
25. Soheilifar MH, Vaseghi H, Seif F, et al. Concomitant overexpression of mir-182-5p and mir-182-3p raises the possibility of IL-17-producing Treg formation in breast cancer by targeting CD3d, ITK, FOXO1, and NFATs: a meta-analysis and experimental study. Cancer Sci. 2021;112(2):589–603. doi:10.1111/cas.14764
26. Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi:10.3322/caac.21660
27. Xu Y, Qiu M, Shen M, et al. The emerging regulatory roles of long non-coding RNAs implicated in cancer metabolism. Mol Ther. 2021;29(7):2209–2218. doi:10.1016/j.ymthe.2021.03.017
28. Ameli-Mojarad M, Ameli-Mojarad M, Nourbakhsh M, Nazemalhosseini-Mojarad E, Silvestris N. Circular RNA hsa_circ_0005046 and hsa_circ_0001791 may become diagnostic biomarkers for breast cancer early detection. J Oncol. 2021;2021:2303946. doi:10.1155/2021/2303946
29. Rao A, Arvinden VR, Ramasamy D, et al. Identification of novel dysregulated circular RNAs in early-stage breast cancer. J Cell Mol Med. 2021;25(8):3912–3921. doi:10.1111/jcmm.16324
30. Wang Y, Li J, Du C, et al. Upregulated circular RNA circ-UBE2D2 predicts poor prognosis and promotes breast cancer progression by sponging miR-1236 and miR-1287. Transl Oncol. 2019;12(10):1305–1313. doi:10.1016/j.tranon.2019.05.016
31. Graffeo R, Rana HQ, Conforti F, et al. Moderate penetrance genes complicate genetic testing for breast cancer diagnosis: ATM, CHEK2, BARD1 and RAD51D. Breast. 2022;65:32–40. doi:10.1016/j.breast.2022.06.003
32. Zeng Y, Zou Y, Gao G, et al. The biogenesis, function and clinical significance of circular RNAs in breast cancer. Cancer Biol Med. 2021;19(1):14–29. doi:10.20892/j.issn.2095-3941.2020.0485
33. Liu Y, Lu C, Zhou Y, Zhang Z, Sun L. Circular RNA hsa_circ_0008039 promotes breast cancer cell proliferation and migration by regulating miR-432-5p/E2F3 axis. Biochem Biophys Res Commun. 2018;502(3):358–363. doi:10.1016/j.bbrc.2018.05.166
34. Nicot C. RNA-seq reveals novel CircRNAs involved in breast cancer progression and patient therapy response. Mol Cancer. 2020;19(1):76. doi:10.1186/s12943-020-01198-2
35. Zhang F, Li L, Fan Z. circRNAs and their relationship with breast cancer: a review. World J Surg Oncol. 2022;20(1):373. doi:10.1186/s12957-022-02842-5
36. Yi Z, Li Y, Wu Y, et al. Circular RNA 0001073 attenuates malignant biological behaviours in breast cancer cell and is delivered by nanoparticles to inhibit mice tumour growth. Onco Targets Ther. 2020;13:6157–6169. doi:10.2147/OTT.S248822
37. Lin X, Chen W, Wei F, Xie X. TV-circRGPD6 nanoparticle suppresses breast cancer stem cell-mediated metastasis via the miR-26b/YAF2 axis. Mol Ther. 2021;29(1):244–262. doi:10.1016/j.ymthe.2020.09.005
38. Chen RX, Liu HL, Yang LL, et al. Circular RNA circRNA_0000285 promotes cervical cancer development by regulating FUS. Eur Rev Med Pharmacol Sci. 2019;23(20):8771–8778. doi:10.26355/eurrev_201910_19271
39. Yuan G, Ding W, Sun B, Zhu L, Gao Y, Chen L. Upregulated circRNA_102231 promotes gastric cancer progression and its clinical significance. Bioengineered. 2021;12(1):4936–4945. doi:10.1080/21655979.2021.1960769
40. Luo ZB, Lai GE, Jiang T, Cao CL, Peng T, Liu FE. A competing endogenous RNA network reveals novel lncRNA, miRNA and mRNA biomarkers with diagnostic and prognostic value for early breast cancer. Technol Cancer Res Treat. 2020;19:1533033820983293. doi:10.1177/1533033820983293
41. Lu JT, Tan CC, Wu XR, et al. FOXF2 deficiency accelerates the visceral metastasis of basal-like breast cancer by unrestrictedly increasing TGF-beta and miR-182-5p. Cell Death Differ. 2020;27(10):2973–2987. doi:10.1038/s41418-020-0555-7
42. Sang Y, Chen B, Song X, et al. circRNA_0025202 regulates tamoxifen sensitivity and tumor progression via regulating the miR-182-5p/FOXO3a axis in breast cancer. Mol Ther. 2019;27(9):1638–1652. doi:10.1016/j.ymthe.2019.05.011
43. Khan MA, Massey S, Ahmad I, et al. FOXO1 gene downregulation and promoter methylation exhibits significant correlation with clinical parameters in Indian breast cancer patients. Front Genet. 2022;13:842943. doi:10.3389/fgene.2022.842943
44. Wu Y, Elshimali Y, Sarkissyan M, Mohamed H, Clayton S, Vadgama JV. Expression of FOXO1 is associated with GATA3 and Annexin-1 and predicts disease-free survival in breast cancer. Am J Cancer Res. 2012;2(1):104–115. doi:10.1245/s10434-009-0481
45. Zeng R, Peng B, Peng E, Chen Z. Downregulated copper homeostasis-related gene FOXO1 as a novel indicator for the prognosis and immune response of breast cancer. J Immunol Res. 2022;2022:9140461. doi:10.1155/2022/9140461
46. Sun WL, He LY, Liang L, et al. Ambra1 regulates apoptosis and chemosensitivity in breast cancer cells through the Akt-FoxO1-Bim pathway. Apoptosis. 2022;27(5–6):329–341. doi:10.1007/s10495-022-01718-z
47. Wu Z, Niu T, Xiao W. Uev1A promotes breast cancer cell survival and chemoresistance through the AKT-FOXO1-BIM pathway. Cancer Cell Int. 2019;19(1):331. doi:10.1186/s12935-019-1050-4
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.