Back to Journals » International Journal of Nanomedicine » Volume 9 » Issue 1
Enhanced cellular uptake of maleimide-modified liposomes via thiol-mediated transport
Received 30 November 2013
Accepted for publication 18 February 2014
Published 5 June 2014 Volume 2014:9(1) Pages 2849—2861
DOI https://doi.org/10.2147/IJN.S58540
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 5
Tianshu Li, Shinji Takeoka
Department of Life Science and Medical Bioscience, Graduate School of Advanced Science and Engineering, Waseda University (TWIns), Tokyo, Japan
Abstract: With a small amount of maleimide modification on the liposome surface, enhanced cellular uptake of liposomes and drug-delivery efficiency can be obtained both in vitro and in vivo. Herein, we describe the mechanisms underlying this enhanced cellular uptake. Suppression of the cellular uptake of maleimide-modified liposomes (M-GGLG, composed of 1,5-dihexadecyl N,N-diglutamyl-lysyl-L-glutamate [GGLG]/cholesterol/poly(ethylene glycol) – 1,2-distearoyl-sn-glycero-3-phosphoethanolamine [PEG5000-DSPE]/maleimide [M]-PEG5000-Glu2C18 at a molar ratio of 5:5:0.03:0.03) caused by temperature block and addition of serum was alleviated compared with that of liposomes without maleimide modification (GGLG liposomes, composed of GGLG/cholesterol/PEG5000-DSPE/PEG5000-Glu2C18 at a molar ratio of 5:5:0.03:0.03). When 0.01 nM N-ethylmaleimide was used to pre-block cellular thiols, the cellular uptake of M-GGLG liposomes was decreased to approximately 70% in HeLa, HCC1954, MDA-MB-468, and COS-7 cell lines. Moreover, inhibition of a thiol-related reductase such as protein disulfide isomerase resulted in a 15%–45% inhibition of the cellular uptake of M-GGLG liposomes, whereas GGLG liposomes were not influenced. Further, single and mixed inhibitors of clathrin-mediated endocytosis, caveolae-mediated endocytosis, and macropinocytosis did not efficiently inhibit the cellular uptake of M-GGLG liposomes. Using confocal microscopy, we verified that M-GGLG liposomes were localized partially in lysosomes after inhibition of the mentioned conventional endocytic pathways. Therefore, it was hypothesized that the mechanisms underlying the enhanced cellular uptake of liposomes by maleimide modification was thiol-mediated membrane trafficking, including endocytosis and energy-independent transport.
Keywords: maleimide, thiol reactive, liposome, endocytosis, energy-independent transport, protein disulfide isomerase
Introduction
Protein oxidations are a major class of protein posttranslational modifications that occur through the reaction between amino acid residues and reactive oxygen species or reactive nitrogen species. The oxidative modification of protein is usually associated with the functional decline or loss of the target protein, and promotes normal aging and age-related pathogenesis. However, it has been now widely and steadily recognized that protein oxidation also plays an indispensable role in cell survival, regeneration, and the recovery of cellular functions such as extension of life span and resistance to severe circumstances.1 Thus, the redox equilibrium is crucial to maintain the regular functions of cell proteins.
Cellular thiols are well known to play an important role in the redox regulation of cellular functions and response to endogenous and exogenous oxidative stress. Many proteins containing cysteine residues in their primary structure are capable of folding and stabilizing their tertiary structure by forming internal disulfide bonds. Particularly in secreted proteins, which encounter strong oxidants in the extracellular milieu, disulfide bonds make a strong contribution to protein stability. Disulfide bonds present in mature proteins can be cleaved into thiols through dithiol–disulfide exchange, alkaline hydrolysis, or acid-based assisted hydrolysis.2 Thioredoxins, which constitute a ubiquitous family of proteins of ~12 kDa with a dithiol/disulfide active site, are the principal cellular protein disulfide reductases in the cytosol, providing electrons for a number of other reductases.3 The protein disulfide isomerase (PDI), a member of the thioredoxin family that catalyzes disulfide formation and isomerization and inhibits aggregation, is responsible for the reduction of disulfide bonds in macromolecular conjugates during endocytosis.4 Other redox enzymes such as gamma-interferon-inducible lysosomal thiol reductase (GILT) also contribute to reduction and redox regulation in lysosomes.5
The reversible nature of disulfide bonds has been exploited in a number of ways for advanced drug delivery via disulfide exchange between the substrate and cellular thiols.6 For example, a protein kinase C peptide inhibitor that is cell impermeant was found to cross cell membranes when an activated cysteine was introduced into its sequence.7 It has been suggested that peptides containing disulfides or thiols can cross-react with cell-surface thiols and then might be either trapped in the membrane or further internalized. Not only peptides, but also other synthetic biomolecules such as oligonucleotides, nanoparticles, polymers, and fluorescent dyes or probes that present thiol-reactive moieties can exhibit enhanced cellular association and internalization.8 However, the exact mechanism of thiol-mediated cellular uptake remains unclear. Thiolated biomolecules might interact with exofacial thiols, followed by standard endocytic pathways to pass through the plasma membrane. There is also another supposition that the thiol-mediated uptake of nanoparticles is independent of endocytic routes that have been extensively investigated such as clathrin-mediated endocytosis (CME), caveolae-mediated endocytosis, and macropinocytosis.9
“Endocytosis” is a fundamental movement in eukaryotic cells involved in nutrient uptake, signal transduction, antigen presentation, neuronal synaptic transmission, transcellular transport, and many other physiological processes.10 During endocytosis, plasma membrane invaginates with the cargoes to form trafficking vehicles, which then break away to transport the membrane-coated vacuoles to organelles or release their contents into cytosol. The structures and functions of a number of coated proteins and adaptor proteins that belong to the network of endocytosis have been investigated by modern molecular and cell biology approaches.11 CME is associated with the assembly of “clathrin”, a coat protein on the intracellular face of the plasma membrane. The cargoes are recruited into clathrin-coated pits, then the local plasma membrane invaginates into cytosol to form the vesicles for intracellular trafficking.12 Pharmacological inhibition of CME involves loss of clathrin and the AP2 adaptor complex, a well-known inhibitor of which is chlorpromazine (CPZ).13 Lipid raft/caveolae-mediated endocytosis indicates the vesicular invagination of cholesterol-enriched microdomains of the cell membrane assisted by a coat protein, namely caveolin. Inhibitors of this pathway may include nystatin (Nys) and methyl-β-cyclodextrin (MβCD), which bind to or deplete cholesterol from the plasma membrane. “Macropinocytosis” is a unique internalization which can be stimulated by mitogens and growth factors, and undergo membrane ruffling to form large vacuoles for the intracellular trafficking of exogenous substances. It is associated with the dynamics of cortical actin and intracellular actin polymerization.14 Cytochalasin D (Cyto D) is often used to depolymerize F-actin by binding to a growing end of its filaments, whereas Wortmannin (Wort) blocks constitutive and stimulated macropinocytosis by inhibiting phosphoinositide 3-kinase.15,16
“Endocytosis” is a fundamental movement in eukaryotic cells involved in nutrient uptake, signal transduction, antigen presentation, neuronal synaptic transmission, transcellular transport, and many other physiological processes.10 During endocytosis, plasma membrane invaginates with the cargoes to form trafficking vehicles, which then break away to transport the membrane-coated vacuoles to organelles or release their contents into cytosol. The structures and functions of a number of coated proteins and adaptor proteins that belong to the network of endocytosis have been investigated by modern molecular and cell biology approaches.11 CME is associated with the assembly of “clathrin”, a coat protein on the intracellular face of the plasma membrane. The cargoes are recruited into clathrin-coated pits, then the local plasma membrane invaginates into cytosol to form the vesicles for intracellular trafficking.12 Pharmacological inhibition of CME involves loss of clathrin and the AP2 adaptor complex, a well-known inhibitor of which is chlorpromazine (CPZ).13 Lipid raft/caveolae-mediated endocytosis indicates the vesicular invagination of cholesterol-enriched microdomains of the cell membrane assisted by a coat protein, namely caveolin. Inhibitors of this pathway may include nystatin (Nys) and methyl-β-cyclodextrin (MβCD), which bind to or deplete cholesterol from the plasma membrane. “Macropinocytosis” is a unique internalization which can be stimulated by mitogens and growth factors, and undergo membrane ruffling to form large vacuoles for the intracellular trafficking of exogenous substances. It is associated with the dynamics of cortical actin and intracellular actin polymerization.14 Cytochalasin D (Cyto D) is often used to depolymerize F-actin by binding to a growing end of its filaments, whereas Wortmannin (Wort) blocks constitutive and stimulated macropinocytosis by inhibiting phosphoinositide 3-kinase.15,16
In our previous research, we developed maleimide modified 1,5-dihexadecyl N,N-diglutamyl-lysyl-L-glutamate (M-GGLG) liposomes by partially functionalizing the PEG layer with maleimide moiety. M-GGLG liposomes showed markedly increased drug-delivery efficiency both in vitro and in vivo in comparison to the unmodified 1,5-dihexadecyl N,N-diglutamyl-lysyl-L-glutamate (GGLG) liposomes.17 Herein, we describe the mechanisms underlying the enhanced cellular uptake of maleimide-modified liposomes, and moreover, investigate the functions of cell-surface thiols during the cellular internalization of thiol-reactive nanoparticles.
Materials and methods
Materials
The following reagents were purchased: maleimide-PEG5000-carboxyl-NHS from NOF Corporation (Tokyo, Japan); 1,2-distearoyl-sn-glycero-3-phosphoethanolamine (DSPE) and cholesterol (chol) from Nippon Fine Chemicals (Osaka, Japan); Alexa Fluor® 488 C5-maleimide from Life Technologies (Carlsbad, CA, USA); CPZ hydrochloride, Nys, MβCD, Cyto D, Wort, and bacitracin from Sigma-Aldrich (St Louis, MO, USA); and rhodamine-PE lipid (Rho-PE) from Avanti Polar Lipids (Alabaster, AL, USA). The required 1,5-dioctadecyl L-glutamate (Glu2C18), PEG5000-Glu2C18, PEG5000-DSPE, GGLG, and maleimide-PEG5000-Glu2C18 were synthesized in our laboratory using previously published methods.17–19
Liposome preparation
M-GGLG liposomes were prepared from GGLG, chol, PEG5000-DSPE, and maleimide-PEG5000-Glu2C18 at a molar ratio of 5:5:0.03:0.03, stirring at room temperature for 4 hours in Dulbecco’s phosphate-buffered saline without magnesium or calcium (DPBS). For preparation of the GGLG liposomes, the lipid PEG5000-Glu2C18 was used instead of maleimide-PEG5000-Glu2C18 at the same molar ratio. After hydration and extrusion, the liposome dispersion was subject to ultracentrifugation at 49,000 rpm for 30 minutes. The supernatant was then removed and the pellet was re-dispersed in DPBS.17 To visualize the liposomes, Rho-PE was added in the initial step at 1 mol% of total lipids. The lipid concentration of the liposomes was calculated from the concentration of chol using a chol kit from Wako Pure Chemical Industries (Osaka, Japan) according to the manufacturer’s instructions.
Cell culture
HeLa, HCC1954, MDA-MB-468, and COS-7 cells were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin. The cells were grown at 37°C in an atmosphere containing 5% CO2, and passaged by trypsinization with 0.1% trypsin-ethylenediaminetetraacetic acid (EDTA).
Influence of temperature block on the cellular uptake of liposomes
Cells were seeded in 24-well plates (5.0 × 104 cells/well) and incubated in an atmosphere of 5% CO2 at 37°C for 24 hours. Next, the cells were precooled at 4°C for 2 hours before exchanging the medium in the cell-culture dish with 400 μL of fresh DMEM containing 72 μg/mL of liposomes. Upon further incubation at 4°C for 2 hours, the cells were washed once with precooled DPBS containing 20 U/mL of heparin sulfate and twice with precooled DPBS, and then 200 μL of 0.5% Triton X-100 buffer was added to lyse the cells. The amount of liposomes internalized in the cells was fluorometrically determined from the lysate using a fluorescence spectrometer (Multi-Detection Microplate Reader, BioTek Instruments, Winooski, VT, USA) with an excitation wavelength of 530 nm and an emission wavelength of 590 nm. The protein concentration of the lysate was determined by a standard protein assay (Pierce 660 nm Protein Assay; Thermo Fisher Scientific, Waltham, MA, USA). The cellular uptake efficiency of the liposomes was expressed as μg of lipid per mg of cellular protein.17
Influence of serum on the cellular uptake of liposomes
Cells were seeded in 24-well plates (5.0 × 104 cells/well) and incubated in an atmosphere of 5% CO2 at 37°C for 24 hours. Next, cells were washed three times with DPBS, and the medium was exchanged for 72 μg/mL of GGLG- or M-GGLG-liposome solution in fresh DMEM or DMEM containing 10% FBS. After incubation at 37°C for 2 hours, the cells were washed once with DPBS containing 20 U/mL of heparin sulfate to remove membrane-bound liposomes and twice with DPBS, then 200 μL of 0.5% Triton X-100 buffer was added to lyse the cells.
Influence of N-ethylmaleimide on the cellular uptake of liposomes
Cells were seeded in 24-well plates (5.0 × 104 cells/well) and incubated in an atmosphere of 5% CO2 at 37°C for 24 hours. To determine the influence of N-ethylmaleimide (NEM), cells were preincubated with 0.01 nM NEM in fresh DMEM for 10 minutes at 37°C under a 5% CO2 atmosphere. The medium in the cell-culture dish was then exchanged with 400 μL of fresh DMEM containing 72 μg/mL of liposomes. Upon incubation at 37°C for 2 hours, the cells were washed once with DPBS containing 20 U/mL of heparin sulfate and twice with DPBS, and then 200 μL of 0.5% Triton X-100 buffer was added to lyse cells.
Study of the endocytic mechanisms of liposomes
Cells were seeded in 24-well plates (5.0 × 104 cells/well) and incubated in an atmosphere of 5% CO2 at 37°C for 24 hours. The medium in the cell-culture dish was then exchanged with 400 μL of fresh DMEM containing 72 μg/mL of liposomes. Upon incubation at 37°C for 2 hours, the cells were washed once with DPBS containing 20 U/mL of heparin sulfate and twice with DPBS, and then 200 μL of 0.5% Triton X-100 buffer was added to lyse the cells. To determine the effect of the inhibition of endocytosis, the cells were preincubated for 30 minutes at 37°C under a 5% CO2 atmosphere with one of the following solutions: 10 μg/mL of CPZ to inhibit CME, 10 μg/mL of Nys or 1 mM MβCD to inhibit caveolae-dependent endocytosis, and 10 μg/mL of Cyto D or 20 μM Wort to inhibit macropinocytosis. For comprehensive inhibition of the endocytic pathways, a mixture of 4 μg/mL of CPZ, 4 μg/mL of Nys, 0.4 mM MβCD, 4 μg/mL of Cyto D, and 5 μM of Wort in DMEM was applied for 30 minutes preincubation with cells at 37°C under a 5% CO2 atmosphere. The relative cellular uptake efficiency was then calculated using the following formula:

Confocal laser scanning microscopy of the intracellular distribution of liposomes
Cells were seeded in six-well glass plates (1.0 × 105 cells/well) and incubated in an atmosphere of 5% CO2 at 37°C for 24 hours. For the inhibition of endocytosis, mixed inhibitors containing 4 μg/mL of CPZ, 4 μg/mL of Nys, 0.4 mM MβCD, 4 μg/mL of Cyto D, and 5 μM of Wort in DMEM were preincubated with cells for 30 minutes at 37°C and then washed with DPBS twice. The culture medium was exchanged for fresh DMEM medium containing 72 μg/mL of M-GGLG or GGLG liposomes and incubated for 30 minutes. Next, the cells were washed once with DPBS containing 20 U/mL of heparin sulfate and twice with DPBS, and then 50 nM LysoTracker® Green DND-26 (Life Technologies) was added for a further 30 minutes’ incubation. Lastly, the cells were washed three times with DPBS, transparent DMEM medium was added, and the cells were observed under a confocal laser scanning microscope (FluoView™ FV1000; Olympus, Tokyo, Japan).
Influence of PDI inhibitors on the cellular uptake of liposomes
Cells were seeded in 24-well plates (5.0 × 104 cells/well) and incubated in an atmosphere of 5% CO2 at 37°C for 24 hours. For inhibition of PDI, cells were preincubated with 1 nM 5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB) or 1 nM bacitracin in DMEM for 30 minutes at 37°C under a 5% CO2 atmosphere. Next, the medium in the cell-culture dish was exchanged with 400 μL of fresh DMEM containing 72 μg/mL of liposomes. Following incubation at 37°C for 2 hours, the cells were washed once with DPBS containing 20 U/mL of heparin and twice with DPBS, then 200 μL of 0.5% Triton X-100 buffer was added to lyse the cells.
Statistical analysis
Student’s t-test was applied to analyze the differences between the M-GGLG liposomes and GGLG liposomes in the experiments studying the influence of various inhibitors on the cellular uptake. A P-value<0.05 was considered statistically significant.
Results
Influence of temperature block on the cellular uptake of liposomes
The entry of nanoparticles into cells is well known to occur via energy-dependent endocytosis10 and/or energy-independent trafficking such as membrane fusion. It was revealed in our previous research that M-GGLG liposomes show higher cellular uptake efficiency than GGLG liposomes at 37°C.17 To study the energy-independent uptake pathway of M-GGLG liposomes, the cellular uptake efficiency was evaluated at a low temperature (ie, 4°C) when most of the energy-dependent activities such as endocytosis were suppressed.
As shown in Table 1, due to temperature block at 4°C, the cellular uptake efficiencies of both GGLG and M-GGLG liposomes were greatly decreased. For GGLG liposomes, the uptake efficiency at 4°C was decreased to 24%–40% of that at 37°C in HeLa, HCC1954, MDA-MB-468, and COS-7 cells. Comparatively, the M-GGLG liposomes exhibited a higher cellular uptake efficiency at 4°C (Figure 1), and the inhibition of cellular uptake by temperature block was also less severe (ie, 35%–67% of the uptake at 37°C; Table 1) in these cell lines. Therefore, it was suggested that the energy-independent transport of liposomes could be enhanced through the surface modification of maleimide.
Influence of serum on the cellular uptake of liposomes
The cooperation of a number of factors and proteins in serum often leads to suppression of the cellular internalization of nanoparticles.20,21 As shown in Figure 2, the cellular uptake efficiencies of both GGLG and M-GGLG liposomes largely declined as a result of serum inhibition in HeLa, HCC1954, MDA-MB-468, and COS-7 cells. The cellular uptake of GGLG liposomes was decreased to 22%–28% of that in serum-free medium. In comparison, the effect of serum inhibition on the cellular uptake of M-GGLG liposomes was weaker in these cell lines; that is, 37%–56% of the cellular uptake efficiency in serum-free medium.
Influence of NEM on the cellular uptake of liposomes
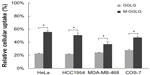
As shown in Figure 3, by pre-blocking cellular thiols with 0.01 nM NEM, the cellular uptake of M-GGLG liposomes was observed to decrease to approximately 70% of the control uptake efficiency in HeLa, HCC1954, MDA-MB-468, and COS-7 cells; in contrast, no significant inhibition of the cellular uptake of GGLG liposomes was observed in these same cell lines.
Study of the endocytic mechanisms of liposomes
Well-known endocytic pathways of nanoparticles include CME, caveolae-dependent endocytosis, and macropinocytosis. Herein, we utilized CPZ to inhibit CME, Nys and MβCD to inhibit caveolae-dependent endocytosis, and Cyto D and Wort to inhibit macropinocytosis.
As shown in Figure 4, the cellular endocytic pathway of liposomes varies according to the type of cell and liposome. For GGLG liposomes, the endocytic rate was reduced to around 50% by each of the inhibitors applied to HeLa, MDA-MB-468, and COS-7 cells, indicating that the endocytic pathways that function in these cell lines include CME, caveolae-dependent endocytosis, and macropinocytosis. In HCC1954 cells, the endocytosis of GGLG liposomes was mainly caveolae dependent, because the cellular uptake was largely inhibited by Nys and MβCD to around 50%. In comparison, the cellular uptake of M-GGLG liposomes was only slightly inhibited by CPZ and/or Nys in HeLa, HCC1954, and COS-7 cells, with the inhibitory rates ranging from 10% to 25% in these cell lines. No significant inhibition of the cellular uptake of M-GGLG liposomes by MβCD, Cyto D, or Wort was observed in any of the cell lines tested. Conversely, the cellular uptake of M-GGLG liposomes was stimulated by MβCD, Cyto D, and Wort in HeLa, MDA-MB-468, and COS-7 cells.
In a separate experiment (shown in Figure 5), a mixture of all the listed endocytosis inhibitors was employed to inhibit the conventional endocytic pathways of liposomes (ie, CME, caveolae-dependent endocytosis, and macropinocytosis) comprehensively. The experimental conditions were the same as those shown in Figure 4 except that the concentration of each inhibitor was decreased (ie, to 40% of the original value) in consideration of the overall cytotoxicity (Figure S1). The cellular uptake efficiency of GGLG liposomes was suppressed to 50%–75% by the mixed inhibitors of conventional endocytic pathways in HeLa, HCC1954, MDA-MB-468, and COS-7 cells. In contrast, the cellular uptake efficiency of M-GGLG liposomes was not significantly inhibited by the mixture of the inhibitors in HeLa, HCC1954, or COS-7 cells; moreover, stimulated internalization was confirmed in MDA-MB-468 cells.
Intracellular distribution of liposomes
In general, the mechanisms underlying the cell-membrane trafficking of nanoparticles can be divided into energy-dependent transport, such as endocytosis, and energy-independent transport, such as membrane fusion.22–24 During endocytosis, coated pits will be formed, which allow the invagination of cell membrane to import macromolecules via endosomes/lysosomes; thus, there is no immediate release of cargoes to the cytosol. In contrast, cell-membrane fusion will directly lead to the translocation of the attached nanoparticles into the cytosol.
In this study, we utilized LysoTracker® to visualize the lysosomes (green dots) and Rho-PE, a component of the liposome bilayer, was applied to trace the location of liposomes (red dots) (Figure 6). As shown in Figure 6C, after cellular internalization without the inhibition of conventional endocytosis, the majority of GGLG liposomes were located in lysosomes and a minority were distributed in the cytosol, revealing that endocytosis was a predominant cellular trafficking pathway for GGLG liposomes. In comparison, a slightly increased distribution of M-GGLG liposomes in the cytosol was observed (red dots in Figure 6I), suggesting that the cellular uptake of these liposomes through energy-independent trafficking was enhanced. However, it should be noted that endocytosis was still a main pathway for the cellular internalization of the M-GGLG liposomes.
As shown in Figure 6F, the employment of mixed inhibitors of endocytosis resulted in a decreased cellular internalization of GGLG liposomes, with no co-localization in lysosomes observed, indicating the complete inhibition of conventional endocytic pathways. The cytoplasmic distribution of GGLG liposomes illustrated that non-endocytic transport occurred. In contrast, the mixed inhibitors of endocytosis did not totally suppress the lysosomal distribution of M-GGLG liposomes (Figure 6L), which suggested the existence of an alternative endocytic pathway for M-GGLG liposomes to enter cells.
Influence of PDI inhibitors on the cellular uptake of liposomes
PDI is a key factor that regulates thiols–disulfides exchange on the cell surface. Well-known inhibitors of PDI include bacitracin and DTNB, which block the reductive functions of this enzyme and are often used as thiol blockers.25,26 As shown in Figure 7, the cellular uptake of M-GGLG liposomes was reduced to 55%–83% of the control group’s by pretreatment with bacitracin or DTNB in HeLa, HCC1954, MDA-MB-468, and COS-7 cells, whereas the uptake of GGLG liposomes was not significantly suppressed by PDI inhibitors.
Discussion
Cell-surface thiols are important functional groups for self-protection against extracellular oxidative stress, and are expressed numerously (Figure S2) and extensively (Figure S3) in different cell lines. It has been reported that cell-surface thiols are involved in enhanced membrane fusion between some viral envelopes and the host-cell membrane during the virus infection process,27,28 and they are highly expressed during tumor cell invasion.29 PDI, a member of the thioredoxin family, is considered one of the key factors regulating these processes. It is also emphasized that the oxidoreductase activity of the PDI superfamily is important in facilitating the cellular uptake of a series of thiol-reactive compounds or particles.8
In our previously disclosed research, maleimide modification at the level of 0.3 mol% of total lipids in the liposomes did not significantly affect their physical characteristics such as size, lipid composition, drug-to-lipid ratio, drug encapsulation efficiency (Table S1), stability, or zeta potential (Figure S4).17 However, the cellular uptake and drug-delivery efficacy of maleimide-modified doxorubicin (DOX)-encapsulating liposomes (M-GGLG-DOX-liposomes) were significantly enhanced both in vitro and in vivo in comparison to the unmodified GGLG-DOX-liposomes. As such, thiol-reactive maleimide moiety was suggested as the key factor in the different biological properties of M-GGLG liposomes. In this research, we investigated the cellular-uptake pathway of M-GGLG liposomes to elucidate the mechanisms underlying thiol-mediated enhanced cellular uptake.
In general, most of the active transport through the cell membrane can be inhibited by temperature block at 4°C.30 Therefore, after washing off any liposomes that might have been attached to the cytoplasm membrane using heparin sulfate, cellular internalization at 4°C indicated the transport of liposomes via an energy-independent mechanism (eg, membrane fusion). As shown in Figure 1 and Table 1, the maleimide modification of the liposomes mitigated the suppression of cellular internalization by temperature block at 4°C, suggesting a relatively enhanced cellular uptake of M-GGLG liposomes via energy-independent transport in comparison with the GGLG liposomes. This is consistent with the observation of the increased distribution of M-GGLG liposomes in cytosol (red dots in Figures 6C and I). Because the reaction of cellular thiols with maleimide can initiate within 15 seconds and reach saturation within 5 minutes at 4°C (Figure S2), it is hypothesized that the enhanced energy-independent trafficking of M-GGLG liposomes was facilitated by the conjugation of maleimide with cellular thiols.
Serum serves as a nutrient source, supplying proteins, carbohydrates, fats, inorganics, and so forth for cell growth and proliferation. These contents enter cells through diffusion, active transport, and endocytosis, and might compete with nanoparticles for entry into the cells. Moreover, serum contains a number of factors that can deactivate or suppress cell-surface proteins/receptors, which lead to the decreased association of nanoparticles with the plasma membrane and subsequent cellular internalization.31,32 Hence, serum has been considered a negative factor during the process of the cellular uptake of nanoparticles. In our study, the maleimide modification of the liposome surface might have triggered an alternative (ie, thiol-mediated) trafficking route, thus the serum suppression of the cellular uptake of M-GGLG liposomes was not as great as that of GGLG liposomes (Figure 2). It is also proposed that the M-GGLG liposomes may have conjugated with some serum proteins (Table S2), such as albumin, which express reductive cysteine residues at the exterior surface and tend to be taken by cells via receptor-mediated endocytosis.33,34 Thus, in contrast with the GGLG liposomes, the cellular internalization of the M-GGLG liposomes may be partially facilitated through conjugation with certain serum proteins. Although, the concentration of cellular thiols could have decreased when the cells were surrounded with serum proteins; there was still a considerable number of thiols that remained active (Figure S5). Therefore, it is surmised that cellular thiols largely compromise the conjugation between serum proteins and M-GGLG liposomes, and that the thiol-mediated transport of M-GGLG liposomes could also occur in 10% serum-containing medium.
“NEM” is a membrane-impermeable compound that can conjugate covalently to thiols. When we utilized a small amount of this low-molecular-weight compound (ie, 0.01 nM) to pre-block active thiol sites on the cell surface, the cellular uptake of M-GGLG liposomes was significantly decreased, whereas that of GGLG liposomes was not (Figure 3). This observation suggested that an interaction between cell-surface thiols and maleimide moiety on liposomes is a prerequisite for the enhanced cellular uptake of M-GGLG liposomes. The recognition of thiols might be involved in the subsequent endocytosis or energy-independent uptake of M-GGLG liposomes. Interestingly, this thiol-mediated internalization might be independent of the conventional endocytosis including CME, caveolae-mediated endocytosis, and macropinocytosis. By employing single or mixed inhibitors of these conventional endocytic routes, the cellular uptake of M-GGLG liposomes was not significantly suppressed (Figures 4 and 5). Moreover, confocal microscopic observation of the intracellular distribution of the M-GGLG liposomes revealed a co-localization of M-GGLG liposomes with lysosomes after the comprehensive inhibition of those conventional endocytic pathways (Figure 6), which also indicated an alternative form of endocytosis for the M-GGLG liposomes. On the other hand, the amount and activity of cell-surface thiols would be maintained and balanced by thioredoxins such as PDI. Without these enzymes, the cell-surface thiols would be easily oxidized by the extracellular environment, resulting in a reduced number of active thiol sites. Therefore, when PDI inhibitors (ie, bacitracin and DTNB) were applied to suppress the cellular thiols, the cellular uptake of the M-GGLG liposomes decreased remarkably (Figure 7), while that of the GGLG liposomes was not interrupted. This phenomenon demonstrated that the reductive function of the thiols on the cell surface is necessary for the enhanced cellular uptake of M-GGLG liposomes. Disulfide reductases such as PDI might be key proteins that perform the recognition of and subsequent alternative endocytosis and energy-independent cellular uptake of thiol-reactive nanoparticles.
Conclusion
In this study, cell-surface thiols facilitated the cellular uptake of maleimide-modified liposomes. The interaction of maleimide with cellular thiols triggered alternative cellular internalization via thiol-mediated transport, including energy-independent membrane trafficking and endocytosis, which is independent of conventional CME, caveolae-mediated endocytosis, and macropinocytosis.
Acknowledgments
This work was partially supported by the Global Center of Excellence (GCOE) “Practical Chemical Wisdom” and “Grants for Excellent Graduate Schools” project of Waseda University from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan. Tianshu Li would also like to thank China Scholarship Council for supporting her PhD study.
Disclosure
The authors declare no conflicts of interest in this work.
References
Cai Z, Yan LJ. Protein oxidative modifications: beneficial roles in disease and health. J Biochem Pharmacol Res. 2013;1(1):15–26. | |
Hogg PJ. Disulfide bonds as switches for protein function. Trends Biochem Sci. 2003;28(4):210–214. | |
Arnér ES, Holmgren A. Physiological functions of thioredoxin and thioredoxin reductase. Eur J Biochem. 2000;267(20):6102–6109. | |
Wilkinson B, Gilbert HF. Protein disulfide isomerase. Biochim Biophys Acta. 2004;1699(1–2):35–44. | |
Arunachalam B, Phan UT, Geuze HJ, Cresswell P. Enzymatic reduction of disulfide bonds in lysosomes: characterization of a gamma-interferon-inducible lysosomal thiol reductase (GILT). Proc Natl Acad Sci U S A. 2000;97(2):745–750. | |
Saito G, Swanson JA, Lee KD. Drug delivery strategy utilizing conjugation via reversible disulfide linkages: role and site of cellular reducing activities. Adv Drug Deli Rev. 2003;55(2):199–215. | |
Aubry S, Burlina F, Dupont E, et al. Cell-surface thiols affect cell entry of disulfide-conjugated peptides. FASEB J. 2009;23(9):2956–2967. | |
Torres AG, Gait MJ. Exploiting cell surface thiols to enhance cellular uptake. Trends Biotechnol. 2012;30(4):185–190. | |
Sahay G, Alakhova DY, Kabanov AV. Endocytosis of nanomedicines. J Control Release. 2010;145(3):182–195. | |
Mukherjee S, Ghosh RN, Maxfield FR. Endocytosis. Physiol Rev. 1997;77(3):759–803. | |
Schmid EM, McMahon HT. Integrating molecular and network biology to decode endocytosis. Nature. 2007;448(7156):883–888. | |
Doherty GJ, McMahon HT. Mechanisms of endocytosis. Annu Rev Biochem. 2009;78:857–902. | |
Ivanov AI. Pharmacological inhibition of endocytic pathways: is it specific enough to be useful? Methods Mol Biol. 2008;440:15–33. | |
Mercer J, Helenius A. Virus entry by macropinocytosis. Nat Cell Biol. 2009;11(5):510–520. | |
Hansen SH, Olsson A, Casanova JE. Wortmannin, an inhibitor of phosphoinositide 3-kinase, inhibits transcytosis in polarized epithelial cells. J Biol Chem. 1995;270(47):28425–28432. | |
Dutta D, Donaldson JG. Search for inhibitors of endocytosis: Intended specificity and unintended consequences. Cell Logis. 2012;2(4):203–208. | |
Li T, Takeoka S. A novel application of maleimide for advanced drug delivery: in vitro and in vivo evaluation of maleimide-modified pH-sensitive liposomes. Int J Nanomedicine. 2013;8:3855–3866. | |
Okamura Y, Maekawa I, Teramura Y, et al. Hemostatic effects of phospholipid vesicles carrying fibrinogen gamma chain dodecapeptide in vitro and in vivo. Bioconjug Chem. 2005;16(6):1589–1596. | |
Obata Y, Tajima S, Takeoka S. Evaluation of pH-responsive liposomes containing amino acid-based zwitterionic lipids for improving intracellular drug delivery in vitro and in vivo. J Control Release. 2010;142(2):267–276. | |
Zelphati O, Uyechi LS, Barron LG, Szoka FC Jr. Effect of serum components on the physico-chemical properties of cationic lipid/oligonucleotide complexes and on their interactions with cells. Biochim Biophys Acta. 1998;1390(2):119–133. | |
Johnstone SA, Masin D, Mayer L, Bally MB. Surface-associated serum proteins inhibit the uptake of phosphatidylserine and poly(ethylene glycol) liposomes by mouse macrophages. Biochim Biophys Acta. 2001;1513(1):25–37. | |
Nel AE, Mädler L, Velegol D, et al. Understanding biophysicochemical interactions at the nano-bio interface. Nat Mater. 2009;8(7):543–557. | |
Verma A, Stellacci F. Effect of surface properties on nanoparticle-cell interactions. Small. 2010;6(1):12–21. | |
Verma A, Uzun O, Hu Y, et al. Surface-structure-regulated cell-membrane penetration by monolayer-protected nanoparticles. Nat Mater. 2008;7(7):588–595. | |
Dickerhof N, Kleffmann T, Jack R, McCormick S. Bacitracin inhibits the reductive activity of protein disulfide isomerase by disulfide bond formation with free cysteines in the substrate-binding domain. FEBS J. 2011;278(12):2034–2043. | |
Mandel R, Ryser HJ, Ghani F, Wu M, Peak D. Inhibition of a reductive function of the plasma membrane by bacitracin and antibodies against protein disulfide-isomerase. Proc Natl Acad Sci U S A. 1993;90(9):4112–4116. | |
Fenouillet E, Barbouche R, Courageot J, Miquelis R. The catalytic activity of protein disulfide isomerase is involved in human immunodeficiency virus envelope-mediated membrane fusion after CD4 cell binding. J Infect Dis. 2001;183(5):744–752. | |
Jain S, McGinnes LW, Morrison TG. Thiol/disulfide exchange is required for membrane fusion directed by the Newcastle disease virus fusion protein. J Virol. 2007;81(5):2328–2339. | |
Goplen D, Wang J, Enger Pø, et al. Protein disulfide isomerase expression is related to the invasive properties of malignant glioma. Cancer Res. 2006;66(20):9895–9902. | |
Willingham MC, Rutherford AV, Gallo MG, et al. Receptor-mediated endocytosis in cultured fibroblasts: cryptic coated pits and the formation of receptosomes. J Histochem Cytochem. 1981;29(9):1003–1013. | |
Smith PJ, Giroud M, Wiggins HL, et al. Cellular entry of nanoparticles via serum sensitive clathrin-mediated endocytosis, and plasma membrane permeabilization. Int J Nanomedicine. 2012;7:2045–2055. | |
Escriou V, Ciolina C, Lacroix F, Byk G, Scherman D, Wils P. Cationic lipid-mediated gene transfer: effect of serum on cellular uptake and intracellular fate of lipopolyamine/DNA complexes. Biochim Biophys Acta. 1998;1368(2):276–288. | |
Schwegler JS, Heppelmann B, Mildenberger S, Silbernagl S. Receptor-mediated endocytosis of albumin in cultured opossum kidney cells: a model for proximal tubular protein reabsorption. Pflugers Arch. 1991;418(4):383–392. | |
Brunskill NJ, Stuart J, Tobin AB, Walls J, Nahorski S. Receptor-mediated endocytosis of albumin by kidney proximal tubule cells is regulated by phosphatidylinositide 3-kinase. J Clin Invest. 1998;101(10):2140–2150. |
Supplementary materials
Characterization of the liposomes
The composition, size distribution and zeta potentials of the liposomes have been shown in Table S1 and Figure S4.
Confocal laser scanning microscopy of thiol distribution on the cell surface
Alexa Fluor® 488 C5-maleimide (Alexa-C5-maleimide), a membrane-impermeable agent that can conjugate covalently to thiols, was used to observe the distribution of thiols on the cell surface. Cells were seeded in six-well glass plates (5 × 104 cells/well) and incubated in an atmosphere of 5% CO2 at 37°C for 24 hours. To prevent the cellular internalization of Alexa-C5-maleimide, cells were precooled on ice for 30 minutes. After washing the cells with precooled Dulbecco’s phosphate-buffered saline (DPBS), 10 μM of Alexa-C5-maleimide in fresh Dulbecco’s Modified Eagle’s Medium (DMEM) without fetal bovine serum (FBS) was added to the cell-culture plates, which were then incubated on ice for 15 seconds, 30 seconds, 1 minute, 2 minutes, 3 minutes, 4 minutes, 5 minutes, 8 minutes and 10 minutes. To verify that the thiol conjugation was irreversible, cells were preincubated with 500 μM N-ethylmaleimide (NEM) for 5 minutes on ice, then washed with precooled DPBS three times, followed by further incubation with 10 μM Alexa-C5-maleimide in fresh DMEM for 5 minutes on ice. Cells were fixed with 4% formalin solution for 15 minutes after washing with precooled DPBS three times, and then observed by confocal laser scanning microscopy.
As shown in Figure S2A, conjugation of maleimide to the cellular thiols was initiated within 15 seconds, and the reaction was completed in about 5 minutes. Figure S2B shows the cell surface dyed with Alexa-C5-maleimide, which disclosed the existence of a large number of free thiols on the cell surface. The distribution of thiols also included cell junctions and adhesions.
Study for the concentration of thiols on different cell surfaces
Cells were seeded in 12-well plates (5 × 104 cells/well) and incubated in an atmosphere of 5% CO2 at 37°C for 24 hours. After cooling the cells on ice for 30 minutes and washing with precooled DPBS, 10 μM of Alexa-C5-maleimide in fresh DMEM was added to the cell-culture plates, which were then incubated for 5 minutes on ice. Next, the cells were washed and lysed with 0.5% Triton X-100, followed by the measurement of the concentration of Alexa-C5-maleimide at an emission wavelength of 528 nm and an excitation wavelength of 485 nm using a fluorescence spectrometer (Multi-Detection Microplate Reader, BioTek Instruments Inc., Winooski, VT, USA). The protein concentration of the lysate was determined by a standard protein assay (Pierce 660 nm Protein Assay, Thermo Fisher Scientific, Waltham, MA, USA). The concentration of cell-surface thiols was represented as nmol of thiols per mg of protein.
Quantitation of the cell-surface thiols was conducted by measuring the fluorescence intensity of Alexa-C5-maleimide bound to the surface thiols. Figure S3 reveals that the concentration of cellular surface thiols varied in different cell lines. HeLa cells showed the highest surface density of thiols among the cell lines tested.
Study for the biocompatibility of endocytosis inhibitors
Figure S1 shows the cytotoxicities of single and mixed endocytosis inhibitors in HeLa, HCC1954, MDA-MB-468 and COS-7 cell lines.
NEM conjugation with free thiols in serum
Heat-inactivated FBS (10%) in DPBS or 0.9 mg/mL bovine serum albumin was incubated with 10 μM Alexa-C5-maleimide at 37°C for 30 minutes. PD-10 desalting columns containing Sephadex® G-25 (GE Healthcare Life Sciences, Little Chalfont, UK) were applied for the separation of protein-conjugated Alexa-C5-maleimide and free Alexa-C5-maleimide. The concentration of Alexa-C5-maleimide was fluorometrically determined using a fluorescence spectrometer (excitation wavelength, 485 nm; emission wavelength, 528 nm) to calculate the ratio of protein-conjugated maleimide to total maleimide, as shown in Table S2.
Influence of serum on the expression of cellular thiols
HeLa cells were seeded in 24-well plates (5.0 × 104 cells/well) and incubated in an atmosphere of 5% CO2 at 37°C for 24 hours. The medium in the cell-culture dish was then exchanged for fresh DMEM containing 0%, 5%, 10%, and 20% FBS (v/v) for a further 24 hours’ incubation. Cells were precooled at 4°C for 2 hours, washed with DPBS thrice, then incubated with 100 μM Alexa-C5-maleimide at 4°C for 10 minutes. Triton X-100 buffer (0.5%) was added to lyse the cells after washing off the Alexa-C5-maleimide. The concentration of Alexa-C5-maleimide was fluorometrically determined from the lysate using a fluorescence spectrometer (excitation wavelength, 485 nm; emission wavelength, 528 nm). The protein concentration of the lysate was determined by a standard protein assay (Pierce 660 nm Protein Assay). The concentration of surface thiols was expressed as mol of thiols per gram of cellular protein. As shown in Figure S5, the expression of cellular thiols was gradually suppressed by increasing the ratio of the serum in the culture medium.
 © 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.