Back to Journals » Drug Design, Development and Therapy » Volume 12
Design, synthesis, docking, and antimicrobial evaluation of some novel pyrazolo[1,5-a]pyrimidines and their corresponding cycloalkane ring-fused derivatives as purine analogs
Authors Abdallah AEM , Elgemeie GH
Received 8 December 2017
Accepted for publication 1 March 2018
Published 20 June 2018 Volume 2018:12 Pages 1785—1798
DOI https://doi.org/10.2147/DDDT.S159310
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Qiongyu Guo
Amira EM Abdallah, Galal H Elgemeie
Department of Chemistry, Faculty of Science, Helwan University, Helwan, Cairo, Egypt
Background: Over the years, pyrazolopyrimidine derivatives have been recognized as having antimicrobial activities. Recently, we reported different synthetic methods to prepare pyrazolopyrimidine derivatives as anticancer and antimicrobial agents. The studies showed that our previously reported 5-aminopyrazoles 2 act as a building block for the preparation of a variety of interesting pyrazolopyrimidines as purine analogs.
Purpose: The objective of this study was to describe the direct new method for preparation of novel pyrazolo[1,5-a]pyrimidine derivatives and their corresponding cycloalkane ring-fused derivatives. Also, the new compounds were tested in vitro for their antibacterial and antifungal activity properties.
Methods: Pyrazolo[1,5-a]pyrimidine derivatives were prepared by the reaction of our previously reported 5-aminopyrazoles 2 with suitable sodium salts of (hydroxymethylene) cycloalkanones and sodium salts of unsaturated ketones.
Results: The structures of the new compounds were characterized according to their mass spectroscopy, 1H NMR, IR and elemental analyses. Compounds 8b, 10e, 10i, and 10n were the most active compounds against Gram-positive and Gram-negative bacterial species. Compound 10i with two moieties of 4-Br-C6H4 revealed increased reactivity compared with ampicillin as standard reference.
Conclusion: About twenty two novel pyrazolo[1,5-a]pyrimidine derivatives and their corresponding cycloalkane ring-fused derivatives were prepared through the reaction of 5-aminopyrazoles 2 with different sodium salts of (hydroxymethylene) cycloalkanones and sodium salts of unsaturated ketones. The antibacterial and antifungal activities of the newly synthesized compounds were evaluated and revealed that compounds 8b, 10e, 10i, and 10n were the most active compounds against Gram-positive and Gram-negative bacterial strains.
Keywords: pyrazolo[1,5-a]pyrimidines, 5-aminopyrazoles, 2-(hydroxymethylene)-1-cycloalkanones, 2-formylcycloalkanones, antibacterial, antifungal, docking studies
Introduction
Antimicrobial resistance threatens prevention and treatment of diseases caused by fungi, bacteria, viruses, and parasites. Diseases caused by infection over time are a growing threat to the overall health of people worldwide. Urgent and preventive action must be taken in all societies.1 Antimicrobial resistance occurs when microscopic organisms such as viruses, bacteria, parasites, and fungi change, when treated with antimicrobial agents such as fungicides, antivirals, and antibiotics. Over time, microorganisms promote and acquire antimicrobial resistance. In consequence, most drugs become virtually ineffective in treatment, and diseases and infections will be persistent in the human body, leading to increased and evolving risk of proliferation in communities and threats to our actual ability to treat infectious and common diseases that are known to lead to death.2 It is clear that antimicrobial resistance occurs naturally and spontaneously over time, usually through genetic changes. Antimicrobial resistance is naturally generated by natural selection from random mutations. When the new gene is made, bacteria can convert genetic information in a horizontal way. If the bacteria carry several resistant genes, they are called multi-resistance bacteria. The effect of antimicrobial resistance is the environmental stress on bacteria, but the mutations that appear in some bacterial cells make them escape the antimicrobial resistance effect. Next, this feature moves to the next offspring, which is characterized as a generation with full antimicrobial resistance. Poor ability to control infection, lack of adequate hygienic conditions, and inadequate proper handling of all types of foods lead to increased prevalence of resistance to all antimicrobials. Patients with an infection caused by drug-resistant bacteria are always at increased risk of poor clinical outcomes and acute death as they consume more medicines and medical resources than other infected patients with non-resistant strains of the same microbes and bacteria.3 Many pyrazolopyrimidines are known to possess antimicrobial and antifungal activities;4 we have recently reported different innovative synthetic methods to prepare pyrazolopyrimidine derivatives that found application and appeared to constitute new classes of anticancer and antimicrobial agents.5,6 A series of one of our previously reported novel 5-aminopyrazoles 17–11 (Figure 1) was used recently by other research groups as a starting material for the construction of pyrazolopyrimidines.12–19 The studies demonstrated that our aminopyrazoles act as a building block for a variety of interesting pyrazolopyrimidines as purine analogs. In another study conducted, our previously reported 5-aminopyrazoles 220,21 (Figure 1) were proven as a good starting synthetic material for the preparation of a variety of interesting pyrazolopyrimidines.22,23 We have reported that 5-aminopyrazoles, aminotriazoles, aminotetrazole, and aminobenzimidazole reacted with sodium salts of (hydroxymethylene) cycloalkanones and sodium salts of unsaturated keto compounds to give the corresponding angular azolopyrimidine derivatives.24–31 These promising results have motivated our research group to continue this work exploring novel molecular mechanisms of these synthetic compounds and their use as chemotherapeutic agents. In view of these findings and as a part of our program directed toward the preparation of potential antimetabolic agents,32 we have recently reported different synthetic methods for the preparation of azoloazines using activated nitriles.33 Many derivatives of these ring systems are considered important as antimetabolites in most biochemical reactions.34 In the light of these reports and in the continuing results of our previous research into the synthesis of biologically active heterocyclic compounds,35 the present research reports a new preparation of cycloalkane ring-fused pyrazolo[1,5-a]pyrimidines 8a–f (Scheme 1) and substituted pyrazolo[1,5-a]pyrimidines 10a–n (Scheme 2) by the reaction of our previously reported 5-aminopyrazole 2 with suitable sodium salts of (hydroxymethylene)-cycloalkanones 7a–d and sodium salts of unsaturated keto compounds 9a–h. The synthesized heterocycles were tested and evaluated for their antifungal and antibacterial activities.
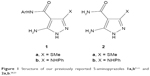  | Figure 1 Structure of our previously reported 5-aminopyrazoles 1a,b7–11 and 2a,b.20,21 |
  | Scheme 1 Synthesis of 7,8-dihydro-6H-cycloalkan[e]pyrazolo[1,5-a]pyrimidine-3-carboxamide derivatives 8a–h. |
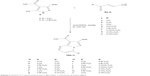  | Scheme 2 Synthesis of 7-substituted-pyrazolo[1,5-a]pyrimidine-3-carboxamide derivatives 10a–n. |
Materials and methods
The melting points were determined on a Gallenkamp melting point apparatus and were uncorrected. Infrared (IR) spectra (KBr discs) were recorded on an Fourier-transform infrared (FTIR) plus 460 IR spectrophotometer (Shimadzu, Japan). 1H NMR spectra were recorded on a BRUKER-400 spectrometer operating at 400 MHz in DMSO-d6 with Si(CH3)4 as an internal standard at the Faculty of Pharmacy, Ain Shams University, Egypt. Shifts were given in ppm and the abbreviations were as follows: s (singlet), d (doublet), t (triplet), and m (multiplet). The mass spectra were run in the Microanalytical Center at Cairo University. The reagents and solvents were purchased in commercially available grade purity. 5-Aminopyrazoles 2 were prepared following our previously reported method.20
Synthetic procedures
General procedure for the synthesis of (8a–h)
To a solution of any of 2b (2.17 g, 0.01 mol) or 2c (2.96 g, 0.01 mol), the sodium salt of 7a (1.34 g, 0.01 mol), 7b (1.48 g, 0.01 mol), 7c (1.62 g, 0.01 mol), or 7d (1.76 g, 0.01 mol) and piperidine acetate (1 mL; prepared from 4.2 mL glacial acetic acid, 10 mL water, and 7.2 mL piperidine) were refluxed in water (50 mL) for 10 min. Acetic acid (1.5 mL) was added to the hot solution and refluxing was continued for about 15 min. The reaction mixture was allowed to cool to room temperature. The precipitate, in each case, was collected by filtration and crystallized from ethanol.
2-(Phenylamino)-7,8-dihydro-6H-cyclopenta[e]pyrazolo[1,5-a]pyrimidine-3-carboxamide (8a)
Canary yellow crystals; yield: 75% (2.21 g), melting point (mp): 310°C–315°C; IR (KBr, υ cm−1): 3,378, 3,301 (NH2), 3,145 (NH), 3,050 (CH-aromatic), 2,955, 2,918 (CH2), 1,651 (C=O), and 1,596, 1,449 (C=C). 1H-NMR (400 MHz DMSO-d6) δ: 2.52–2.50 (m, 2H, CH2), 3.06–3.33 (m, 4H, 2CH2), 6.94–7.36, 7.68–7.70 (m, 5H, C6H5), 7.48–7.57 (s, 2H, NH2), 8.54 (s, 1H, CH pyrimidine), 9.69 (s, 1H, NH). analysis calculated (Anal. Calcd.) for C16H15N5O (293.32): C, 65.52; H, 5.15; N, 23.88. Found: C, 65.69; H, 4.90; N, 24.01.
2-((4-Bromophenyl)amino)-7,8-dihydro-6H-cyclopenta[e]pyrazolo-[1,5-a]pyrimidine-3-carboxamide (8b)
Faint yellow crystals; yield: 80% (2.96), mp: 282°C–290°C; IR (KBr, υ cm−1): 3,405, 3,266 (NH2), 3,168 (NH), 3,040 (CH-aromatic), 2,954, 2,855 (CH2), 1,651 (C=O) and 1,593, 1,450 (C=C). 1H-NMR (DMSO-d6) δ: 2.17–2.26 (m, 2H, CH2), 2.93–3.04 (m, 4H, 2CH2), 7.45–7.49 (s, 2H, NH2), 7.60–7.67 (m, 4H, C6H4), 8.52 (s, 1H, CH pyrimidine), 9.75 (s, 1H, NH). Anal. Calcd. for C16H14N5OBr (372.22): C, 51.63; H, 3.79; N, 18.82. Found: C, 51.89; H, 4.01; N, 18.99.
2-(Phenylamino)-6,7,8,9-tetrahydropyrazolo[1,5-a]quinazoline-3-carboxamide (8c)
White crystals; yield: 99% (3.04 g), mp: 263°C–266°C; IR (KBr, υ cm−1): 3,393, 3,306 (NH2), 3,184 (NH), 3,050 (CH-aromatic), 2,936, 2,862 (CH2), 1,650 (C=O) and 1,595, 1,455 (C=C). 1H-NMR (DMSO-d6) δ: 1.77–1.94 (m, 4H, 2CH2), 2.75–3.11 (m, 4H, 2CH2), 6.94–7.36, 7.65–7.71 (m, 5H, C6H5), 7.51–7.53 (s, 2H, NH2), 8.87 (s, 1H, CH pyrimidine), 9.59 (s, 1H, NH). Anal. Calcd. for C17H17N5O (307.35): C, 66.43; H, 5.58; N, 22.79. Found: C, 66.77; H, 5.31; N, 22.98.
2-((4-Bromophenyl)amino)-6,7,8,9-tetrahydropyrazolo[1,5-a]quinazoline-3-carboxamide (8d)
Off white crystals; yield: 96% (3.70 g), mp: 280°C–285°C; IR (KBr υ cm−1): 3,393, 3,392 (NH2), 3,272 (NH), 3,159 (CH-aromatic), 2,933 (CH2), 1,650 (C=O) and 1,592, 1,453 (C=C). 1H-NMR (DMSO-d6) δ: 1.73–1.88 (m, 4H, 2CH2), 2.51–2.98 (m, 4H, 2CH2), 7.28 (s, 2H, NH2), 7.44–7.60 (m, 4H, C6H4), 8.74 (s, 1H, CH pyrimidine), 9.61 (s, 1H, NH). Anal. Calcd. for C17H16N5OBr (386.25): C, 52.86; H, 4.18; N, 18.13. Found: C, 53.02; H, 4.01; N, 18.40.
2-(Phenylamino)-6,7,8,9,10-pentahydropyrazolo[1,5-a]quinazoline-3-carboxamide (8e)
White crystals; yield: 70% (2.24 g), mp: 222°C–224°C; IR (KBr, υ cm−1): 3,350, 3,300 (NH2, NH), 3,020 (CH-aromatic), 2,960 (CH2), 1,670 (C=O). 1H-NMR (DMSO-d6) δ: 1.60–2.00 (m, 6H, 3CH2), 2.65–3.20 (m, 4H, 2CH2), 6.90–7.80 (m, 5H, C6H5), 7.80–7.83 (s, 2H, NH2), 8.89 (s, 1H, CH pyrimidine), 10.11 (s, 1H, NH). Anal. Calcd. for C18H19N5O (321.38): C, 67.27; H, 5.96; N, 21.79. Found: C, 67.00; H, 5.70; N, 22.00.
2-((4-Bromophenyl)amino)-6,7,8,9,10-pentahydropyrazolo[1,5-a]quinazoline-3-carboxamide (8f)
White powder; yield: 60% (2.40 g), mp: 241°C–245°C; IR (KBr υ cm−1): 3,400, 3,300 (NH2, NH), 3,130 (CH-aromatic), 2,900 (CH2), 1,660 (C=O). 1H-NMR (DMSO-d6) δ: 1.37–1.90 (m, 6H, 3CH2), 2.40–2.90 (m, 4H, 2CH2), 7.00 (s, 2H, NH2), 7.20–7.90 (m, 4H, C6H4), 8.50 (s, 1H, CH pyrimidine), 9.22 (s, 1H, NH). Anal. Calcd. for C18H18N5OBr (400.27): C, 54.01; H, 4.53; N, 17.50. Found: C, 54.00; H, 4.22; N, 17.20.
2-(Phenylamino)-6,7,8,9,10,11-hexahydrocycloocta[e]pyrazolo[1,5-a]pyrimidine-3-carboxamide (8g)
White crystals; yield: 80% (2.69 g), mp: 244°C–245°C; IR (KBr, υ cm−1): 3,397, 3,324 (NH2), 3,273 (NH), 3,146 (CH-aromatic), 2,920, 2,853 (CH2), 1,655 (C=O) and 1,596, 1,447 (C=C). 1H-NMR (DMSO-d6) δ: 1.34–1.44 (m, 4H, 2CH2), 1.69–1.89 (m, 4H, 2CH2), 2.50–3.38 (m, 4H, 2CH2), 6.94–7.55 (m, 5H, C6H5), 7.68–7.70 (s, 2H, NH2), 8.46 (s, 1H, CH pyrimidine), 9.63 (s, 1H, NH). Anal. Calcd. for C19H21N5O (335.40): C, 68.04; H, 6.31; N, 20.88. Found: C, 68.29; H, 6.52; N, 21.10.
2-((4-Bromophenyl)amino)-6,7,8,9,10,11-hexahydrocycloocta[e]pyrazolo[1,5-a]pyrimidine-3-carboxamide (8h)
Faint yellow crystals; yield: 90% (3.70 g), mp: 266°C–270°C; IR (KBr, υ cm−1): 3,367, 3,303 (NH2), 3,269 (NH), 3,157 (CH-aromatic), 2,921, 2,851 (CH2), 1,648 (C=O) and 1,591, 1,453 (C=C). 1H-NMR (DMSO-d6) δ: 1.33–1.42 (m, 4H, 2CH2), 1.67–1.87 (m, 4H, 2CH2), 2.51–2.97 (m, 4H, 2CH2), 7.38–7.58 (m, 4H, C6H4), 7.65–7.67 (s, br, 2H, NH2), 8.44 (s, 1H, CH pyrimidine), 9.68 (s, 1H, NH). Anal. Calcd. for C19H20N5OBr (414.30): C, 55.08; H, 4.87; N, 16.90. Found: C, 55.22; H, 4.61; N, 17.10.
General procedure for the synthesis of (10a–n)
To a mixture of any of 2b (2.17 g, 0.01 mol) or 2c (2.96 g, 0.01 mol), the sodium salt of 9a (1.08 g, 0.01 mol), 9b (1.22 g, 0.01 mol), or 9c (1.70 g, 0.01 mol), 9d (2.04 g, 0.01 mol), 9e (2.49 g, 0.01 mol), 9f (2.00 g, 0.01 mol), 9g (1.84 g, 0.01 mol), or 9h (1.86 g, 0.01 mol) and piperidine acetate (1 mL) were refluxed in water (50 mL) for 10 min. Acetic acid (1.5 mL) was added to the hot solution and refluxing was continued for about 15 min. The reaction mixture was allowed to cool to room temperature. The precipitate, in each case, was collected by filtration and crystallized from ethanol.
7-Methyl-2-(phenylamino)pyrazolo[1,5-a]pyrimidine-3-carboxamide (10a)
Faint brown crystals; yield: 65% (1.74 g), mp: 248°C–250°C; IR (KBr, υ cm−1): 3,395, 3,271 (NH2), 3,161 (NH), 3,050 (CH-aromatic), 2,900 (CH3), 1,651 (C=O) and 1,594, 1,447 (C=C). 1H-NMR (DMSO-d6) δ: 2.52–2.78 (s, 3H, CH3), 7.38–7.58 (m, 5H, C6H5), 7.72–7.74 (s, br, 2H, NH2), 8.52 (s, 1H, CH pyrimidine), 8.98 (s, 1H, CH pyrimidine), 9.67 (s, 1H, NH). Anal. Calcd. for C14H13N5O (267.29): C, 62.91; H, 4.90; N, 26.20. Found: C, 63.03; H, 4.71; N, 26.40.
7-Ethyl-2-(phenylamino)pyrazolo[1,5-a]pyrimidine-3-carboxamide (10b)
Off white crystals; yield: 78% (2.21 g), mp: 215°C–219°C; IR (KBr, υ cm−1): 3,395, 3,273 (NH2), 3,166 (NH), 3,040 (CH-aromatic), 2,925 (CH2, CH3), 1,653 (C=O), and 1,595, 1,449 (C=C). 1H-NMR (DMSO-d6) δ: 1.35–1.39 (t, 3H, CH3), 3.11–3.17 (q, 2H, CH2), 6.95–7.37, 7.60–7.71 (m, 5H, C6H5), 7.44–7.52 (s, br, 2H, NH2), 8.52 (s, 1H, CH pyrimidine), 8.83 (s, 1H, CH pyrimidine), 9.59 (s, 1H, NH). Anal. Calcd. for C15H15N5O (281.31): C, 64.04; H, 5.37; N, 24.90. Found: C, 64.33; H, 5.61; N, 25.20.
2-((4-Bromophenyl)amino)-7-ethylpyrazolo[1,5-a]pyrimidine-3-carboxamide (10c)
Faint yellow crystals; yield: 97% (3.48 g), mp: 217°C–220°C; IR (KBr, υ cm−1): 3,423, 3,315 (NH2), 3,264–3,178 (NH), 3,012 (CH-aromatic), 2,973, 2,919 (CH2, CH3), 1,659 (C=O) and 1,591, 1,455 (C=C). 1H-NMR (DMSO-d6) δ: 1.37–1.41 (t, 3H, CH3), 3.14–3.20 (q, 2H, CH2), 7.06–7.51, 7.63–7.71 (m, 4H, C6H4), 7.57 (s, br, 2H, NH2), 8.58 (s, 1H, CH pyrimidine), 8.91 (s, 1H, CH pyrimidine), 9.63 (s, 1H, NH). Anal. Calcd. for C15H14N5OBr (360.21): C, 50.02; H, 3.92; N, 19.44. Found: C, 50.22; H, 4.02; N, 19.20.
7-Phenyl-2-(phenylamino)pyrazolo[1,5-a]pyrimidine-3-carboxamide (10d)
Canary yellow crystals; yield: 97% (3.51 g), mp: 237°C–240°C; IR (KBr, υ cm−1): 3,376, 3,312 (NH2), 3,269 (NH), 3,149 (CH-aromatic), 1,654 (C=O) and 1,596, 1,444 (C=C). 1H-NMR (DMSO-d6) δ: 6.92–7.67 (m, 10H, 2C6H5), 8.19–8.20 (s, 2H, NH2), 8.65 (s, 1H, CH pyrimidine), 8.66 (s, 1H, CH pyrimidine), 9.66 (s, 1H, NH). Anal. Calcd. for C19H15N5O (329.36): C, 69.29; H, 4.59; N, 21.26. Found: C, 69.45; H, 4.70; N, 21.44.
2-((4-Bromophenyl)amino)-7-phenylpyrazolo[1,5-a]pyrimidine-3-carboxamide (10e)
Canary yellow crystals; yield: 73% (2.96 g), mp: 287°C–290°C; IR (KBr, υ cm−1): 3,412, 3,310 (NH2), 3,265 (NH), 3,166 (CH-aromatic), 1,658 (C=O) and 1,596, 1,455 (C=C). 1H-NMR (DMSO-d6) δ: 7.35–7.69 (m, 9H, C6H5, C6H4), 8.21 (s, 2H, NH2), 8.69 (s, 1H, CH pyrimidine), 8.70 (s, 1H, CH pyrimidine), 9.71 (s, 1H, NH). Anal. Calcd. for C19H14N5OBr (408.25): C, 55.90; H, 3.46; N, 17.15. Found: C, 56.10; H, 3.80; N, 17.30.
7-(4-Chlorophenyl)-2-(phenylamino)pyrazolo[1,5-a]pyrimidine-3-carboxamide (10f)
Canary yellow crystals; yield: 96% (3.47 g), mp: 272°C–275°C; IR (KBr, υ cm−1): 3,381, 3,317 (NH2), 3,271 (NH), 3,169 (CH-aromatic), 1,659 (C=O) and 1,597, 1,452 (C=C). 1H-NMR (DMSO-d6) δ: 6.94–7.78 (m, 9H, C6H5, C6H4), 8.26–8.82 (s, 2H, NH2), 8.34 (s, 1H, CH pyrimidine), 8.69 (s, 1H, CH pyrimidine), 9.75 (s, 1H, NH). MS (EI): m/z (%) 366 [M+2]+ (4.49), 365 [M+1]+ (20.13), 364 [M+] (13.72), 363 [M-1]+ (58.89), 362 [M-2]+ (1.09), 346 (100.00). Anal. Calcd. for C19H14N5OCl (363.80): C, 62.73; H, 3.88; N, 19.25. Found: C, 62.99; H, 4.01; N, 19.40.
2-((4-Bromophenyl)amino)-7-(4-chlorophenyl)pyrazolo[1,5-a]pyrimidine-3-carboxamide (10g)
Canary yellow crystals; yield: 94% (4.14 g), mp: 272°C–275°C; IR (KBr, υ cm−1): 3,409, 3,220 (NH2, NH), 3,050 (CH-aromatic), 1,636 (C=O) and 1,586, 1,468 (C=C). 1H-NMR (DMSO-d6) δ: 6.14–7.94 (m, 8H, 2C6H4), 7.97–7.99 (s, 2H, NH2), 8.11 (s, 1H, CH pyrimidine), 8.25 (s, 1H, CH pyrimidine), 9.72 (s, 1H, NH). Anal. Calcd. for C19H13N5OBrCl (442.70): C, 51.55; H, 2.96; N, 15.82. Found: C, 51.69; H, 3.01; N, 15.97.
7-(4-Bromophenyl)-2-(phenylamino)pyrazolo[1,5-a]pyrimidine-3-carboxamide (10h)
Canary yellow crystals; yield: 99% (4.04 g), mp: 275°C–278°C; IR (KBr, υ cm−1): 3,400, 3,378 (NH2), 3,271 (NH), 3,168 (CH-aromatic), 1,656 (C=O) and 1,596, 1,450 (C=C). 1H-NMR (DMSO-d6) δ: 6.94–7.92 (m, 9H, C6H5, C6H4), 8.18–8.20 (s, 2H, NH2), 8.69 (s, 1H, CH pyrimidine), 8.70 (s, 1H, CH pyrimidine), 9.67 (s, 1H, NH). MS (EI): m/z (%) 410 [M+2]+ (13.48), 409 [M+1]+ (61.29), 408 [M+] (15.20), 407 [M-1]+ (60.21), 406 [M-2]+ (1.25), 390 (100.00). Anal. Calcd. for C19H14N5OBr (408.25): C, 55.90; H, 3.46; N, 17.15. Found: C, 56.09; H, 3.78; N, 17.35.
7-(4-Bromophenyl)-2-((4-bromophenyl)amino)pyrazolo[1,5-a]pyrimidine-3-carboxamide (10i)
Yellow crystals; yield: 85% (4.14 g), mp: 197°C–200°C; IR (KBr, υ cm−1): 3,406, 3,350 (NH2), 3,270 (NH), 3,169 (CH-aromatic), 1,635 (C=O) and 1,588, 1,468 (C=C). 1H-NMR (DMSO-d6) δ: 6.13–7.79 (m, 8H, 2C6H4), 7.89–7.92 (s, 2H, NH2), 8.70 (s, 1H, CH pyrimidine), 8.81 (s, 1H, CH pyrimidine), 9.73 (s, 1H, NH). Anal. Calcd. for C19H13N5OBr2 (487.15): C, 46.84; H, 2.69; N, 14.38. Found: C, 47.02; H, 2.99; N, 14.60.
7-(4-Methoxyphenyl)-2-(phenylamino)pyrazolo[1,5-a]pyrimidine-3-carboxamide (10j)
Canary yellow crystals; yield: 75% (2.69 g), mp: 247°C–250°C; IR (KBr, υ cm−1): 3,362, 3,310 (NH2), 3,167 (NH), 3,040 (CH-aromatic), 1,658 (C=O) and 1,599, 1,456 (C=C). 1H-NMR (DMSO-d6) δ: 3.91 (s, 3H, CH3), 6.94–7.66 (m, 9H, C6H5, C6H4), 8.26–8.28 (s, 2H, NH2), 8.62 (s, 1H, CH pyrimidine), 8.90 (s, 1H, CH pyrimidine), 9.66 (s, 1H, NH). MS (EI): m/z (%) 361 [M+2]+ (2.43), 360 [M+1]+ (17.62), 359 [M+] (74.11), 358 [M-1]+ (2.12), 357 [M-2]+ (0.07), 342 (100.00). Anal. Calcd. for C20H17N5O2 (359.38): C, 66.84; H, 4.77; N, 19.49. Found: C, 67.02; H, 4.99; N, 19.60.
2-((4-Bromophenyl)amino)-7-(4-methoxyphenyl)pyrazolo[1,5-a]pyrimidine-3-carboxamide (10k)
Yellow crystals; yield: 95% (4.14 g), mp: 217°C–220°C; IR (KBr, υ cm−1): 3,407, 3,277 (NH2), 3,155 (NH), 3,050 (CH-aromatic), 1,682 (C=O) and 1,596, 1,461 (C=C). 1H-NMR (DMSO-d6) δ: 3.92 (s, 3H, CH3), 6.10–7.65 (m, 8H, 2C6H5), 8.25–8.27 (s, 2H, NH2), 8.62 (s, 1H, CH pyrimidine), 8.73 (s, 1H, CH pyrimidine), 9.72 (s, 1H, NH). Anal. Calcd. for C20H16N5O2Br (438.28): C, 54.81; H, 3.68; N, 15.98. Found: C, 55.10; H, 3.98; N, 16.11.
2-(Phenylamino)-7-(p-tolyl)pyrazolo[1,5-a]pyrimidine-3-carboxamide (10l)
Canary yellow crystals; yield: 76% (2.60 g), mp: 257°C–260°C; IR (KBr, υ cm−1): 3,372, 3,312 (NH2), 3,270 (NH), 3,156 (CH-aromatic), 1,653 (C=O) and 1,599, 1,450 (C=C). 1H-NMR (DMSO-d6) δ: 2.46 (s, 3H, CH3), 6.93–7.65 (m, 9H, C6H5, C6H4), 8.14–8.16 (s, 2H, NH2), 8.64 (s, 1H, CH pyrimidine), 8.65 (s, 1H, CH pyrimidine), 9.66 (s, 1H, NH). MS (EI): m/z (%) 345 [M+2]+ (2.17), 344 [M+1]+ (17.27), 343 [M+] (70.81), 342 [M-1]+ (1.89), 341 [M-2]+ (0.12), 325 (100.00). Anal. Calcd. for C20H17N5O (343.38): C, 69.96; H, 4.99; N, 20.40. Found: C, 70.21; H, 4.67; N, 20.73.
2-((4-Bromophenyl)amino)-7-(p-tolyl)pyrazolo[1,5-a]pyrimidine-3-carboxamide (10m)
Yellow crystals; yield: 99% (4.20 g), mp: 217°C–220°C; IR (KBr, υ cm−1): 3,400, 3,315 (NH2), 3,260 (NH), 3,156 (CH-aromatic), 1,644 (C=O) and 1,590, 1,470 (C=C). 1H-NMR (DMSO-d6) δ: 2.46 (s, 3H, CH3), 6.11–7.63 (m, 8H, 2C6H4), 8.12–8.14 (s, 2H, NH2), 8.64 (s, 1H, CH pyrimidine), 8.66 (s, 1H, CH pyrimidine), 9.70 (s, 1H, NH). Anal. Calcd. for C20H16N5OBr (422.28): C, 56.89; H, 3.82; N, 16.58. Found: C, 57.14; H, 4.02; N, 16.70.
7-(2-Hydroxyphenyl)-2-(phenylamino)pyrazolo[1,5-a]pyrimidine-3-carboxamide (10n)
Faint brown crystals; yield: 65% (2.26 g), mp: 207°C–210°C; IR (KBr, υ cm−1): 3,442, 3,415 (NH2), 3,351 (NH), 3,159 (CH-aromatic), 1,662 (C=O) and 1,583, 1,447 (C=C). 1H-NMR (DMSO-d6) δ: 6.70–7.20 (m, 9H, C6H4, C6H5), 7.32–7.33 (s, 2H, NH2), 8.88 (s, 1H, CH pyrimidine), 8.99 (s, 1H, CH pyrimidine), 8.98 (s, 1H, NH), 11.01 (s, 1H, OH). Anal. Calcd. for C19H15N5O2 (345.35): C, 66.08; H, 4.38; N, 20.28. Found: C, 66.22; H, 4.01; N, 20.45.
Docking studies and structure–activity relationship
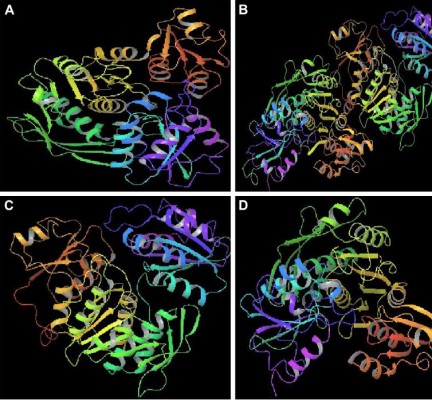
In the absence of a crystal structure, homology models of Bacillus subtilis (Bsu) MurC (Accession No:NP_390857), Pseudomonas aeruginosa (Pae) MurC (Accession No: B7UZI9), and Staphylococcus aureus (Sau) MurC (Accession No:A6U2K6) were built based on the published cocrystal structure of Escherichia coli (Eco) MurC (pdb ID: 2F00; Figure 2). The sequence identities between Eco MurC and Bst MurC, Pae MurC, and Sau MurC were found to be 26%, 58%, and 25%, respectively. The E. coli ATP binding site (126GTHGKTT132) was conserved by >85% with B. subtilis (108GAHGKTSTT116), P. aeruginosa (122GTHGKTT128), and S. aureus (108GAHGKTSTT116). The model was validated using protein Preparation Wizard and minimized prior to docking. Docking of a set of pyrazolopyrimidines was carried out in the ATP binding site using Schrodinger 16.4 software Glide (XP) extra precision module from Schrodinger.36,37 The best Docking Score is obtained as the most negative value for the active ligands. All the compounds were constructed using the fragment library of Maestro 9.2, and all compounds were prepared by using the LigPrep 2.9.38 Glide docking parameters were set to the default hard potential function. No constraints were applied for all the docking studies. Structure–activity relationship (SAR) analysis was performed using R-Group Analysis.
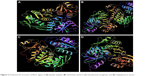  | Figure 2 Predicted 3D structure of MurC ligase of (A) Bacillus subtitles, (B) Escherichia coli K12, (C) Pseudomonas aeruginosa, and (D) Staphylococcus aureus. |
Antimicrobial evaluation
The antimicrobial activities of the samples tested were studied on Mueller–Hinton agar plates by the disc diffusion technique against Gram-positive (B. subtilis and S. aureus) and Gram-negative (E. coli and P. aeruginosa) bacterial strain.39 Ampicillin (AM 20 μg) was used as the standard antibacterial agent obtained from Bioanalyse® Ltd. (Ankara, Turkey). Sterile Whatman filter paper discs (6 mm) were individually impregnated with 10 μL of solvent (distilled water, chloroform, DMSO) containing 20 μg concentration of each sample at a pH value of 6. All the discs were dried aseptically and placed on the surface of Mueller–Hinton agar plates seeded with 1.8×108 cfu/mL (0.5 OD600) of the test bacteria. Following 24 h incubation at 37°C, plates were examined for the presence of inhibition zones. The inhibition zones surrounding the disks were measured (mm) considering only halos >6 mm.40 Inhibition zones obtained are the mean of three replicates for each experiment.
Results and discussion
Chemistry
In this study, 5-aminopyrazoles 2b was found to react with the sodium salts of (hydroxymethylene)-cycloalkanones 7a–d in acetic acid-piperidine acetate to give adduct for which structure 8a–h is set. The reaction starts with an initial nucleophilic attack from the external amino group to the formyl group followed by cyclization and then removal of one molecule of water to produce angular three-ring compounds 8a–h. This requires that in the presence of acidic medium, it occurs by first protonation of the ring nitrogen which is the most nucleophilic center in the compound 2 and directs the exocyclic amino group to attack the unhindered formyl group of 4 to give compounds 8a–h (Scheme 1). The structures of later compounds were confirmed by the spectral data and elemental analysis. Thus, the IR spectrum of compound 8a, as an example of this series, revealed the presence of three bands at υ 3,378, 3,301, and 3,145 cm−1 for NH2 and NH groups and a characteristic C=O band at υ 1,651 cm−1. Moreover, 1H NMR of 8a showed the existence of a signal at δ 8.54 ppm assigned for pyrimidine-H proton, two multiplets at a range of δ 2.52–3.33 ppm assigned for three CH2 groups, and two broad singlets at δ 7.48–7.57 and 9.69 ppm assigned for NH2 and NH groups. The behavior of the 5-aminopyrazoles 2b toward sodium salts of unsaturated keto compounds 9a–h was also studied: the pyrazolopyrimidine compounds 10a–n were obtained by cyclic condensation of 2b with 9a–h in acetic acid-piperidine acetate (Scheme 2). The structure of the 10a–n reaction products was confirmed by spectral data and elemental analysis (IR, 1H NMR, MS). Thus, analytical data were revealed for 10a molecular formula C14H13N5O (M+ 267). The IR spectrum of compound 10a revealed the presence of three bands at υ 3,395, 3,271, and 3,161 cm−1 for NH2 and NH groups and a characteristic C=O band at υ 1,651 cm−1. Also, 1H NMR revealed a multiplet at δ 7.38–7.58 ppm assignable to the aromatic protons and two signals at δ 8.52 and 8.98 ppm assignable for two pyrimidine CH protons. These results obtained in this study, when combined with our previous results, show that the reaction of 5-aminopyrazoles with sodium salts of (hydroxymethylene)-cycloalkanones and sodium salts of unsaturated aliphatic ketones can be used as a new and effective method in the preparation of many important pyrazolo[1,5-a]pyrimidine derivatives and their cycloalkane ring-fused derivatives.
Antimicrobial evaluation and the structure–activity relationship
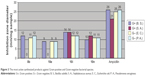
Mur ligases play a vital role in the bio-bacterial peptidoglycans.41,42 Mur ligases play an important role in the biosynthesis of the cell wall peptidoglycan. Many enzymes stimulate the early stages of the pathogenesis of the peptidoglycan named MurA to MurF.43 MurC, the third enzyme in Mur ligases of the peptidoglycan pathway, initiates the synthesis of pentapeptide precursor where the L-alanine binds to the UDP-N-acetylmuramic acid converting to UDP-N-acetylmuramic acid-L-alanine.42 The reactivity of all the newly synthesized products against bacterial and fungi species was evaluated through Table 1. Pyrazolopyrimidine compounds possess bactericidal activity against both Gram-negative and Gram-positive MurC enzymes.42 Compounds 8b, 10e, 10i, and 10n were found to be the most active compounds against Gram-positive and Gram-negative bacterial species (Figure 3). The presence of the two moieties of 4-Br-C6H4 in compound 10i increased the reactivity of the compound when comparing with ampicillin as a standard reference.
  | Table 1 Reactivity of the newly synthesized products against bacterial and fungi species |
The effects of substituents (R1, R2, and R3) in the pyrazolopyrimidine-3-carboxamide region were investigated, and a series of derivatives are summarized in Table 2 and Figure 4. Compound 7-(4-bromophenyl)-pyrazolopyrimidine-3-carboxamide (10i) with 17-Br and 13-bromophenyl substituents exhibited most inhibitory effect for MurC ligase of Gram-negative and Gram-positive bacteria (Table 2). To investigate the binding mode of 10i, it was docked into the active site of MurC ligase of Bacillus subtilis. As shown in Figure 5A, compound 10i interacted with extended conformation. The bromophenyl group formed face-to-face π–π interactions with His263. The carbonyl oxygen atom formed hydrogen bonding interaction with a side chain of Ser275. However, compound 10i formed hydrogen bonding interaction with Gly205 in the case of S. aureus (Figure 5B). MurC ligase of E. coli formed hydrogen bonding interaction with Asn 194 (Figure 5C). P. aeruginosa MurC ligase formed 3 H-bonding interactions with Gln 318, Gln 325, and Val 326 (Figure 5D). However, 7-(4-bromophenyl)-pyrazolopyrimidine-3-carboxamide (10h) showed only inhibitory activities against Bacillus subtilis.7-(2-hydroxyphenyl)-pyrazolopyrimidine-3-carboxamide (10n) containing 2-hydroxyphenyl possessed good inhibitory activities for all MurC ligases of Gram-positive and Gram-negative bacteria (Table 2). Compounds bearing R3 Br-substituents of the 2-(phenylamino)-pyrazolopyrimidine-3-carboxamide displayed better potency for the MurC ligase than those without substituents at the same positions. However, introducing Br-substituents to 2-((4-bromophenyl)amino)-7-(p-tolyl)pyrazolopyrimidine-3-carboxamide (10m) removed its inhibitory activities in comparison with 2-(phenylamino)-7-(4-methylphenyl)pyrazolopyrimidine-3-carboxamide (10l; Table 2). Compounds 8c and 8g did not show any inhibitory activities for MurC ligase of Gram-negative and Gram-positive bacteria (Table 2). However, Br-substitution of compounds 8d and 8h evidently increased their inhibitory effects for MurC ligase (Table 2).
  | Table 2 SAR activates for MurC ligase of 2-(phenylamino)pyrazolo[1,5-a]pyrimidine-3-carboxamide moiety |
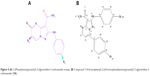  | Figure 4 (A) 2-(Phenylamino)pyrazolo[1,5-a]pyrimidine-3-carboxamide moiety. (B) Compound 7-(4-bromophenyl)-2-((4-bromophenyl)amino)pyrazole[1,5-a]pyrimidine-3-carboxamide (10i). |
  | Figure 5 Binding mode analysis of (10i) with MurC ligase (A) Bacillus subtilis, (B) Escherichia coli K12, (C) Pseudomonas aeruginosa, and (D) Staphylococcus aureus. |
Conclusion
The conclusion of this study was summarized through the reaction of 5-aminopyrazoles 2 with different sodium salts of (hydroxymethylene) cycloalkanones and sodium salts of unsaturated ketones to obtain the novel pyrazolo[1,5-a]pyrimidine derivatives and their corresponding cycloalkane ring-fused derivatives. The newly synthesized compounds were evaluated according to their antibacterial and antifungal activities. The evaluations showed that compounds 8b, 10e, 10i, and 10n were the most active compounds against Gram-positive and Gram-negative bacterial strains.
Author contributions
GHE and AEMA conceived, designed, and performed the experiments; GHE and AEMA analyzed the data, contributed reagents/materials/analysis tools; GHE and AEMA wrote and approved the final manuscript.
Disclosure
The authors report no conflicts of interest in this work.
References
Cassir N, Rolain JM, Brouqui P. A new strategy to fight antimicrobial resistance: the revival of old antibiotics. Front Microbiol.2014;5:551. | ||
Goossens H, Ferech M, Vander Stichele R, Elseviers M. Outpatient antibiotic use in Europe and association with resistance: a cross-national database study. Lancet. 2005;365(9459):579–587. | ||
Mishra RP, Oviedo-Orta E, Prachi P, Rappuoli R, Bagnoli F. Vaccines and antibiotic resistance. Curr Opin Microbiol.2012;15(5):596–602. | ||
Khobragade CN, Bodade RG, Konda SG, Dawane BS, Manwar AV. Synthesis and antimicrobial activity of novel pyrazolo[3,4-d]pyrimidin derivatives. Eur J Med Chem.2010;45(4):1635–1638. | ||
Elgemeie GH, Abu-Zaied MA, Loutfy SA. 4-Aminoantipyrine in carbohydrate research: design, synthesis and anticancer activity of a novel class of derivatives of 4-aminoantipyrine thioglycosides and their corresponding pyrazolopyrimidine and pyrazolopyridine thioglycosides. Tetrahedron. 2107;73:5853–5861. | ||
Elgemeie GH, Abu-Zaied M, Hebishy A, Abbas N, Hamed M. A first microwave-assisted synthesis of a new class of purine and guanine thioglycoside analogs. Nucleosides Nucleotides Nucleic Acids. 2016;35(9):459–478. | ||
Elgemeie GH, Elghandour AH, Elzanate AM, Ahmed SA. Synthesis of some novel α-cyanoketene S,S-acetals and their use in heterocyclic synthesis. J Chem Soc Perkin Trans 1. 1997;(21):3285–3290. | ||
Elgemeie GH, Elghandour AH, Abd-Elaziz GW. Potassium 2-cyanoethylene-1-thiolate derivative: a new preparative route to 2-cyanoketene S,N-acetals and pyrazole derivatives. Synth Commun. 2004;34(18):3281–3291. | ||
Elgemeie GH, Jones PG. 5-Amino-3-anilino-N-(chlorophenyl)-1H-pyrazole-4-carboxamide ethanol solvent. Acta Cryst. 2004;E60:01616–01617. | ||
Kurz T, Widyan K, Elgemeie GH, Novel synthesis of fluorinated cyanoketene N,S-acetals (IV) and their conversions to fluorinated pyrazole derivatives (V). Phosphorus Sulfur Silicon Relat Elem. 2006;181(2):299–304. | ||
Elgemeie GH, Zaghary WA, Amin KM, Nasr TM. A direct route to a new class of acrylamide thioglycosides. J Carbohydr Chem. 2008;27(6):373–378. | ||
Ahmed SA, Hussein AM, Hozayen WGM, El-Ghandour AHH, Abdelhamid AO. Synthesis of some pyrazolopyrimidines as purine analogues. J Heterocyclic Chem. 2007;44(4):803–810. | ||
Ahmed OM, Mohamed MA, Ahmed RR, Ahmed SA. Synthesis and anti-tumor activities of some new pyridines and pyrazolo[1,5-a]pyrimidines. Eur J Med Chem. 2009;44(9):3519–3523. | ||
Ammar YA, Aly MM, Al-Sehemi AG, Salem MA, El-Gaby MSA. Cyanoacetanilides intermediates in heterocyclic synthesis. part 5: preparation of hitherto unknown 5-aminopyrazole and pyrazolo[1,5-a]pyrimidine derivatives containing sulfamoyl moiety. J Chin Chem Soc. 2009;56:1064–1071. | ||
Hussein AM. Synthesis of some new purine-related compounds: regioselective one-pot synthesis of new tetrazolo[1,5-a]pyrimidine, pyrazolo[1,5-a]pyrimidine and pyrimido[1,6-a]pyrimidine derivatives. J Saudi Chem Soc. 2010;14(1):61–68. | ||
Hussein AM. Novel synthesis of some new pyrimido[1,6-a]pyrimidines and pyrazolo[1,5-a]pyrimidine derivatives. J Heterocyclic Chem. 2012;49(2):446–451. | ||
Hassan AS, Hafez TS, Osman SAM, Ali MM. Synthesis and in vitro cytotoxic activity of novel pyrazolo[1,5-a]pyrimidines and related Schiff bases. Turk J Chem. 2015;39:1102–1113. | ||
Hassan AS, Hafez TS, Osman SA. Synthesis, characterization, and cytotoxicity of some new 5-aminopyrazole and pyrazolo[1,5-a]pyrimidine derivatives. Sci Pharm. 2015;83(1):27–39. | ||
Hassan AS, Mady MF, Awad HM, Hafez TS. Synthesis and antitumor activity of some new pyrazolo[1,5-a]pyrimidines. Chin Chem Lett. 2017;28:388–393. | ||
Elgemeie GH, Elghandour AH, Abd Elaziz GW. Novel cyanoketene N,S-acetals and pyrazole derivatives using potassium 2-cyanoethylene-1-thiolates. Synth Commun. 2007;37(17):2827–2834. | ||
Elgemeie GH, Elsayed SH, Hassan AS. Direct route to a new class of acrylamide thioglycosides and their conversions to pyrazole derivatives. Synth Commun. 2008;38(16):2700–2706. | ||
Hafez TS, Osman SA, Yosef HAA, et al. Synthesis, structural elucidation, and in vitro antitumor activities of some pyrazolopyrimidines and Schiff bases derived from 5-amino-3-(arylamino)-1H-pyrazole-4-carboxamides. Sci Pharm. 2013;81(2):339–358. | ||
Hassan AS, Moustafa GO, Awad HM. Synthesis and in vitro anticancer activity of pyrazolo[1,5-a]pyrimidines and pyrazolo[3,4-d][1,2,3]triazines. Synth Commun. 2017;47(21):1963–1972. | ||
Elgemeie GH, Ali HA. Potential purine analogue antagonists: synthesis of novel cycloalkane ring-fused pyrazolo[1,5-a]pyrimidines. Synth Commun. 2002;32(2):253–264. | ||
Elgemeie GH, Hani AA, Jones PG. 2-Phenyl-7,8-dihydro-6H-cyclopenta[e]pyrazolo[1,5-a]pyrimidine. Acta Cryst. 2002;E58:1247–1249. | ||
Elgemeie GH, Helal MH, Ahmed KA. Synthesis and dyeing properties of a new class of condensed carbocyclic arylazopyrazolo[1,5-a]pyrimidines. Pigm Resin Technol. 2003;32(1):10–23. | ||
Elgemeie GH, Fathy NM, Farag DS. Antimetabolites: a novel synthesis of nonclassical condensed carbocyclic purine analogues. Egypt J Pharm Sci. 1997;38(4–6):351–361. | ||
Elgemeie GH, Metwally NH. Synthesis of structurally related purines: benzimidazo[1,2-a]pyridines, benzimidazo[1,2-c]pyrimidines, and pyrazolo[1,5-a]pyrimidines. Monatsh Chem. 2000;131:779–785. | ||
Elgemeie GH, Fathy NM, Hopf H, Jones PG. 1,2,3,4-Tetrahydrobenzimidazo[2,1-b]quinazoline. Acta Cryst. 1998;C54(8):1109–1111. | ||
Elgemeie GEH, Fathy NM, Hopf H, Dix I, Jones PG. 2,3,4,5-Tetrahydrocyclohepta[b]pyrido[2,1-b]benzimidazole-7-carbonitrile. Acta Cryst. 1998;C54:1107–1108. | ||
Elgemeie GEH, Metwally NH, Jones PG. 1,2-Dimethylpyrido[1,2-a]benzimidazole-4-carbonitrile. Acta Cryst. 1998;C54:1871–1873. | ||
Elgemeie GH, El-Ezbawy SR, Ali HA. Reactions of chlorocarbonyl isocyanate with 5-aminopyrazoles and active methylene nitriles: a novel synthesis of pyrazolo[1,5-a]-1,3,5-triazines and barbiturates. Synth Commun. 2001;31(22):3459–3467. | ||
Elgemeie GH, Abu-Zaied MA. Heterocyclic thioglycosides in carbohydrate research: synthesis of thiophene thioglycosides. Nucleosides NucleotidesNucleic Acids. 2017;36(8):511–519. | ||
Elgemeie GH, Fathy NM, Farag AB, Al-Kursani SA. Antimetabolites: design, synthesis, and cytotoxic evaluation of novel dihydropyridine thioglycosides and pyridine thioglycosides. Nucleosides Nucleotides Nucleic Acids. 2017;36(5):355–377. | ||
Elgemeie GH, Jones PG. Crystal structure of 1-amino-2-oxo-2,5,6,7,8,9-hexahydro-1H-cyclohepta[b]pyridine-3-carbonitrile. Acta cryst. 2016;E72:1239–1240. | ||
Small-Molecule Drug Discovery Suite. Glid. version 6.1, New York, NY, USA: Schrödinger, LLC; 2013:20. | ||
Friesner RA, Murphy RB, Repasky MP, et al. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J Med Chem. 2006;49(21):6177–6196. | ||
Release S. 1: LigPrep, version 2.9. New York, NY, USA: Schrödinger, LLC; 2014. | ||
Bauer AW, Kirby WM, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1966;45(4):493–496. | ||
Lapenda JC, Silva Pa, Vicalvi MC, Sena KXFR, Nascimento SC. Antimicrobial activity of prodigiosin isolated from Serratia marcescens UFPEDA 398. World J Microbiol Biotechnol. 2014;31(2):399–406. | ||
Humnabadkar V, Prabhakar KR, Narayan A, et al. UDP-N-acetylmuramic acid l-alanine ligase (MurC) inhibition in a tolC mutant Escherichia coli strain leads to cell death. Antimicrob Agents Chemother. 2014;58(10):6165–6171. | ||
Hameed PS, Manjrekar P, Chinnapattu M, et al. Pyrazolopyrimidines establish MurC as a vulnerable target in Pseudomonas aeruginosa and Escherichia coli. ACS Chem Biol. 2014;9(10):2274–2282. | ||
Kahne D, Leimkuhler C, Lu W, Walsh C. Glycopeptide and lipoglycopeptide antibiotics. Chem Rev. 2005;105(2):425–448. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.