Back to Journals » Chronic Wound Care Management and Research » Volume 10
Ability of Different Non-Antimicrobial Wound Dressings to Remove Bacteria from Surfaces Using in vitro Planktonic and Mature Biofilm Models
Authors Meredith K, Jones AA, Towers VL, Metcalf DG
Received 14 June 2023
Accepted for publication 28 August 2023
Published 5 September 2023 Volume 2023:10 Pages 1—9
DOI https://doi.org/10.2147/CWCMR.S421986
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Marco Romanelli
Kate Meredith,1 Alison Amanda Jones,1 Victoria Louise Towers,1 Daniel Gary Metcalf2
1Microbiology R&D, Convatec Limited, Deeside, Flintshire; 2Advanced Wound Care R&D, Convatec Limited, Deeside, Flintshire
Correspondence: Kate Meredith, Microbiology R&D, Convatec Limited, GDC First Avenue, Deeside Industrial Park, Deeside, Flintshire, Tel +44 7921620140, Email [email protected]
Introduction: Non-antimicrobial wound dressings can remove bacteria from wound surfaces through mechanisms such as binding and immobilization, which may contribute to antimicrobial stewardship.
Methods: This study evaluated four different types of dressings (gauze, carboxymethylcellulose gelling fiber [CMC], dialkylcarbamoyl chloride [DACC] hydrophobic coated fibers, and polyurethane [PU] foam) for removal of planktonic bacteria (all dressings) and mature biofilm bacteria (CMC, DACC and PU foam dressings) in vitro. Total viable counts were performed after incubation for 2, 4 and 6 hours.
Results: The percentage of CA-MRSA removed by CMC dressing was significantly (p< 0.05) greater than all other dressings at all timepoints in the planktonic and biofilm models. A significantly greater percentage of planktonic ESBL P. aeruginosa was removed by CMC dressings than other test dressings with the exception of when compared to PU Foam at the 6-hour time point. Differences in the removal of ESBL P. aeruginosa biofilms between CMC dressings and other dressings were less pronounced, which is likely due to the nature of the biofilm formed.
Conclusion: CMC dressings may play an effective role in reducing bioburden of acute and hard-to-heal wounds in a clinical setting.
Keywords: antimicrobial stewardship, biofilms, microbial drug resistance, wound healing, wound infection
Introduction
Microorganism cell surfaces vary between species and within species depending on genetics, leading to differences in how they attach to surfaces.1 Microorganisms such as bacteria, yeast and moulds exist in two different phenotypes, which affect how they behave and replicate. Planktonic microorganisms consist of single, isolated cells that are unattached to each other and have no need to communicate with each other. They have a high metabolism and multiply rapidly, but are isolated, so are easier to kill with effective antimicrobials. Biofilm is a more complex structure of microbes that attach to each other or to a surface, such as a wound bed or wound tissues. The surface of bacterial cells aids in attachment, which shifts their gene expression, and leads to the beginning of the more complex phenotype, biofilm.2,3 Biofilm usually contains multiple species of microorganisms that can communicate with each other and are encased in a matrix of extracellular polymeric substances, which provides a significant structural defence.4 Biofilm microorganisms also have slower replication and metabolic rates compared with their planktonic counterparts. These attributes mean that biofilm is much more difficult to control and eradicate than planktonic microorganisms.5 Both states may impair healing6 and cause infection in both acute and hard-to-heal wounds,7 however, biofilm is considered a key barrier to wound healing and is present in the majority of hard-to-heal wounds.8
There are a variety of options to reduce bioburden in wounds, including administration of antibiotics, wound cleansers, physical and chemical debridement, and wound dressings, which may or may not contain antiseptics or anti-biofilm technologies. Treatment must be tailored according to the clinical characteristics of the wound and be continually assessed and adapted as the wound moves through the healing trajectory.
Dressings can help to reduce bioburden in the wound, with many containing antiseptics such as iodine, silver or polyhexamethylene biguanide (PHMB).9 Such dressings have been shown to kill microorganisms in in vitro studies, ex vivo evaluations, and in the clinic.9,10 However, balanced against the need to manage microbial bioburden in wounds, there are increasing advances in antimicrobial stewardship,11 where a step-up/step-down approach has been advocated so that antimicrobial dressings are used only when necessary.12 Therefore, other dressings that lack antimicrobial agents, but instead use physical methods such as binding, adsorption, immobilization or sequestration to remove microbes, also play a part in the overall wound management program, when used as part of a protocol of care.
There are many types of gelling fiber dressings manufactured for wound care, such as alginates, chitosan, carboxymethylcellulose (CMC) and polyvinyl alcohol (PVA), which, upon absorption of wound fluid, swell and form a gel.13 CMC gelling fiber dressing has been shown, using scanning electron microscopy in vitro, to effectively encapsulate large populations (up to 70%) of potentially pathogenic bacteria such as Pseudomonas aeruginosa and Staphylococcus aureus. Fewer bacteria were immobilized within the gel matrix of hydrated alginate wound dressings, which appeared to be because they did not form a uniform, cohesive gel structure.14 Staining of bacteria to visualize their viability indicated that the CMC dressing did not kill bacteria; however, it did immobilize them, which prevented them from multiplying.15 Foam dressings, including polyurethane (PU) foam dressings, are widely used to help with the management of exudate.16 Although their ability to absorb fluid has been studied widely, their ability to remove bacteria has not, as the dressing’s focus is usually as a barrier against external bacteria.
Another technology that uses a physical mode of action to attract bacteria is dressings coated with a hydrophobic fatty acid derivative, dialkylcarbamoyl chloride (DACC). The bacterial cell surface contains both hydrophilic and hydrophobic components. These hydrophobic components on the bacterial cell surface contribute to interactions with host cells and other surfaces to initiate colonisation, biofilm formation and infection.17 The hydrophobic properties of DACC have been shown to bind bacteria in both planktonic (Staphylococci) and biofilm (P. aeruginosa and methicillin-resistant S. aureus [MRSA]) forms.17,18 P. aeruginosa was shown to be bound for up to 20 hours without replicating.17
The aim of this study was to compare four different types of non-antimicrobial dressings in their ability and capacity to remove antibiotic-resistant planktonic bacteria and mature biofilm bacteria using in vitro models over short periods of time. The dressings tested were a gauze, a CMC gelling fiber dressing, a hydrophobic DACC-coated dressing, and a PU foam dressing. These are four different dressing technologies which are known to differ in their fluid handling capabilities,19 but to the best of our knowledge, their ability to remove planktonic and biofilm bacteria has not been compared.
Materials and Methods
Test Dressings
Four dressings were tested to evaluate their ability to remove bacteria from planktonic models:
- A sterile knitted viscose gauze dressing (N-A® Gauze, 3M, UK) (Gauze)
- A CMC gelling fiber dressing (Aquacel® Extra™ dressing; Convatec Ltd, UK) (CMC)
- A dressing coated with DACC (Sorbact® Compress; ABIGO Medical AB, Sweden) (DACC)
- A PU foam dressing (Mepilex®; Mölnlycke Health Care Limited, UK) (PU foam).
The CMC, DACC and PU foam dressings were also tested to evaluate their ability to remove bacteria from biofilm models.
Preparation of Inoculated Surface Model (Planktonic Bacteria)
Separate suspensions of each challenge organism, extended-spectrum beta-lactamase (ESBL) P. aeruginosa (NCTC 13437) and community-acquired MRSA (CA-MRSA) (ATTC® BAA™-1556, clone of USA300), were inoculated to a final concentration of approximately 5×106 colony forming units (CFU)/mL in simulated wound fluid (SWF, maximum recovery diluent [MRD; Lab M, UK]/Foetal Bovine Serum [FBS; Biowest, France] in 50:50 v/v). Whatman Grade 1 filter discs (sterile, 42.5 mm diameter) were placed onto the center of pre-dried 90 mm Tryptone Soy Agar (TSA; Lab M, UK) plates (to represent the nutritious wound bed environment) and inoculated with a 200 μL volume of the challenge organism suspension.
Preparation of Mature Biofilm Model
Separate suspensions of each challenge organism, ESBL P. aeruginosa (NCTC 13437) and CA-MRSA (ATTC® BAA™-1556), were inoculated into Tryptone Soy Broth (LAB M, UK)/FBS (50:50 v/v) to give a final concentration of approximately 1×106 CFU/mL. N-A® Gauze samples, 44 mm in diameter (the substrate for biofilm development), were added to the challenge organism suspensions and incubated at 35±3°C for 48 hours in a shaking incubator. Following incubation, gauze-biofilm samples were washed in 0.85% saline (Oxoid, Ireland), to remove planktonic and loosely adhered bacteria. Gauze-biofilm samples required for testing were transferred to the center of a TSA plate (to represent the nutritious wound bed environment). A total viable count was performed on the control gauze-biofilm samples by placing them into a stomacher bag containing 30 mL MRD containing 0.01% Tween 80 (Fisher Bioreagents, US). BacLight® Live/Dead™ stain (Molecular Probes, Invitrogen, US) was used to stain the gauze-biofilm samples to visualize the bacteria within the biofilm. The BacLight® Live/Dead™ stain was prepared as described by the manufacturer and then added in a 200 µL volume to a 1×1 cm section of the gauze-biofilm sample. This was left in darkness for 10 minutes, then confocal laser scanning microscopy (CLSM; LSM800 with Airyscan, Zeiss, Germany) was used to image the biofilm using ×40 objective lens with 488 nm and 630 nm lasers to excite the fluorophores within the BacLight® Live/Dead™ stain.
Evaluation of Bacterial Removal by Test Dressings Using Planktonic and Biofilm Models
Test dressing samples (5×5 cm portions) were applied on top of the inoculated filter discs/gauze-biofilms. To ensure maximum dressing contact between dressing and filter disc/gauze-biofilms, a metal disc (approximately 3 cm in diameter and 8 g in weight) was applied centrally over the dressings prior to incubation. The test samples were incubated at room temperature for 2, 4 and 6 hours. Short durations were used to examine the binding ability of the dressings, which avoided the potential of bacterial multiplication which would happen over longer timepoints and affect results. Five repeats were prepared for each test dressing, at each time point and for both challenge organisms. Filter disc/gauze-biofilm controls without dressings were assessed at 0, 2, 4 and 6 hours for each organism, ensuring that total numbers of the challenge organisms were monitored throughout testing.
Total Viable Counts for Planktonic and Biofilm Models
Following incubation, each dressing was transferred to a stomacher bag containing a 40 mL volume of MRD containing 0.01% Tween 80 (Fisher, UK). All samples were homogenized in the stomacher for 4 minutes to loosen and remove the bacteria from the dressings (historical usage of this method has shown effective recovery of bacteria from dressing materials within this laboratory; data not shown). Counts were performed on all test set-ups. For filter disc/gauze-biofilm following dressing application and for the no-dressing filter disc/gauze-biofilm controls, the total viable counts were performed as above but in 30 mL of MRD containing 0.01% Tween 80. To confirm differences between the dressings (for both planktonic and biofilm data), statistical analysis was performed where results for CFU/filter, CFU/gauze or CFU/dressing, and percentage of challenge organism removed into each dressing ([CFU recovered from dressing ÷ total CFU [the amount recovered from the dressing and the remaining numbers on the surface, ie, filter or gauze]] × 100), were compared for significant differences at each time point for each organism using one-way analysis of variance (ANOVA) and Tukey grouping. Results with an ANOVA p value of <0.05 and sample means in different Tukey groups (illustrated with different symbols per timepoint) were considered to be significantly different. If test dressing columns have the same symbol within that timepoint, then they were not classed as significantly different.
Results
Removal of Planktonic ESBL P. aeruginosa and CA-MRSA
Total viable counts remained high for the no-dressing filter disc control throughout the 6-hour test period for both ESBL P. aeruginosa (≥2.5×105 CFU/filter disc) and CA-MRSA (≥3.1×105 CFU/filter disc). The mean percentage of ESBL P. aeruginosa and CA-MRSA taken up by the test dressings compared with the total amount present is illustrated in Figure 1. Comparing the mean ranges over the testing periods for ESBL P. aeruginosa, the CMC dressing removed 41–58% of the total amount of bacteria compared with 12–21% for the gauze dressing, 4–8% for the DACC dressing and 12–27% for the PU foam dressing. An even greater difference was observed for CA-MRSA cells, where the CMC dressing removed 77–79% of total bacteria compared with 20–25% for the gauze dressing, <1% for the DACC dressing and 10–15% for the PU foam dressing. The differences in ability to remove bacteria between the CMC dressing and the other test dressings were shown to be statistically significant (p<0.05 and a different Tukey group to the other test dressings) for planktonic CA-MRSA following 2, 4 and 6 hours contact with the inoculated surface; this was also observed with planktonic ESBL P. aeruginosa, with the exception of CMC dressings compared to PU Foam at the 6-hour time point (Figure 1).
Binding of ESBL P. aeruginosa and CA-MRSA Mature Biofilms
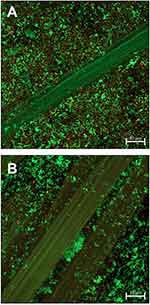
Visualization of the ESBL P. aeruginosa and CA-MRSA biofilms on the gauze substrate stained with BacLight™ Live/Dead® using CLSM confirmed the presence and viability of mature biofilm (Figures 2A and B). Initial biofilm counts for ESBL P. aeruginosa and CA-MRSA were approximately 1.5×1010 and 2.7×1010 CFU/gauze-biofilm, respectively. Total viable counts for the no-dressing gauze-biofilm control remained high for both ESBL P. aeruginosa (≥4.8×109CFU/gauze-biofilm) and CA-MRSA (≥8.4×108 CFU/gauze-biofilm) throughout the 6-hour test period. The mean percentage of ESBL P. aeruginosa and CA-MRSA biofilm cells taken up by the test dressings compared with the total amount of bacteria present is illustrated in Figure 3, with statistical difference illustrated in the graph. Comparing the mean ranges over the testing periods for ESBL P. aeruginosa, the CMC dressing removed 58–60% of the total amount of bacteria compared with 15–39% for the DACC dressing and 26–39% for the PU foam dressing. In general, a lower percentage of MRSA biofilm was removed by the test dressings, however there was still a significantly greater proportion removed by the CMC dressing (16–42% of total bacteria) than the DACC dressing (<1%) and the PU foam dressing (approximately 2%). These differences in ability to remove biofilm between the CMC and the other dressings were shown to be significant (p<0.05 and a different Tukey group to other dressings) for CA-MRSA following 2, 4 and 6 hours contact with the biofilm (Figure 3). In contrast, due to result variation leading to larger error bars, CMC dressing only removed significantly more ESBL P. aeruginosa than the DACC dressing and PU foam dressing at 4 hours (p-<0.05) (Figure 3).
Discussion
The dressings tested were able to remove varying degrees of planktonic and biofilm bacteria from surfaces, which interestingly, did not appear to be related to fluid handling capabilities. For example, foam dressings tend to have high fluid handling capacity19 yet this did not aid bacterial removal by the PU foam dressing under test. The CMC dressing physically removed more planktonic and mature biofilm bacteria (ESBL P. aeruginosa and CA-MRSA) than the gauze, DACC and PU foam dressings tested in this study. These differences were all statistically significant for the CMC dressing, with the exception of planktonic ESBL P. aeruginosa at 6 hours and ESBL P. aeruginosa biofilm at 2 and 6 hours.
The removal of planktonic ESBL P. aeruginosa and CA-MRSA in the current study are consistent with observations by Walker et al who also demonstrated that up to 70% of planktonic pathogens (S. aureus and P. aeruginosa) are retained in CMC dressings through sequestration.14 A previous in vitro study by Bowler et al, compared the ability of CMC, DACC and two alginate dressings (Algosteril®, Les Laboratoires Brothier, and Kaltostat®, Convatec) to sequester and retain planktonic bacteria.13 The CMC dressing and the DACC dressing both effectively retained 60–80% of S. aureus and P. aeruginosa, whereas the two alginate dressings retained only 10% of S. aureus and 30–40% of P. aeruginosa. The retention of bacteria by the DACC dressing in this earlier study was much higher than observed in the current study, which is likely a result of the different testing methods used. The previous study used a model that evaluated the uptake of bacteria from a solution, whereas the current study assessed the uptake of bacteria when attached to a surface, which is more likely to represent how bacteria would be present in a real wound environment.
In the planktonic model in this current study, filter discs were used to contain the inoculated challenge organism, keeping it in a particular area of the TSA to ensure that the dressing came into contact with all bacteria present. Filters are commonly used in microbiology when performing solution sampling to ascertain how many bacteria are present,20 or as a substrate for biofilm growth.21 In common with these tests, in this study a nutrient agar was placed under the filter enabling the bacteria to be supplied with nutrients and replicate whilst being immobilized on the filter. It is therefore not unexpected that the total viable counts remained high for the no-dressing filter disc control throughout the 6-hour test period for both test species. Confirmation of the suitability of gauze as a substrate for biofilm growth was shown by visualization of the biofilm using CLSM, where mature viable biofilm was observed. In addition, total viable counts remained high for both test organisms during the 6-hour test period, ensuring that the biofilm bacteria remained viable throughout the testing period.
In the current study, the CMC dressing removed a similar percentage of the total amount present for ESBL P. aeruginosa regardless of whether it was in planktonic or mature biofilm phenotype. In contrast, the amount of CA-MRSA removed by the CMC dressing was markedly influenced by the bacterial phenotype, with far greater amounts of planktonic bacteria removed compared to the mature biofilm phenotype. Although the difference between bacterial phenotypes was more apparent for CA-MRSA for all dressings, the results obtained for ESBL P. aeruginosa showed greater variation, as shown by the larger standard deviations, suggesting that all the dressings performed inconsistently against biofilms for both challenge organisms. This is likely due to the extracellular polymeric substance (EPS) produced by the bacterial cells within the biofilms, protecting the bacterial cells and reducing their removal. It was also observed that more biofilm bacteria were removed by all dressings for ESBL P. aeruginosa compared with MRSA, with DACC and PU foam dressings barely removing any CA-MRSA biofilm bacteria, which could also be an indication of how the dressings are able to bind different types of biofilms. It is widely known that ESBL P. aeruginosa biofilms are more mucoid, due to the production of alginate in the EPS,22,23 than CA-MRSA biofilms; this was visually apparent during this testing (data not shown). Previous studies have also shown differences between S. aureus/MRSA and P. aeruginosa biofilms, showing MRSA with cells packed close together, forming dense biofilms and P. aeruginosa less close to each other.24 Therefore, these physical biofilm differences may have caused the differences in bacteria/biofilm uptake observed between dressings in this study.
DACC is a hydrophobic dressing, which was exhibited via the difference in bacteria removal between the challenge organisms in this study. DACC removed planktonic and biofilm ESBL P. aeruginosa bacteria more successfully when compared with CA-MRSA. This is likely due to the nature of the cell surface of CA-MRSA; the production of carbohydrate polymer and teichoic acid in their cell wall has been shown to decrease their removal by hydrophobic dressings.17
Retention of bacteria within a wound dressing could raise concerns regarding potential risk of infection. However, a previous in vitro laboratory study that stained bacteria to visualize their viability found that, although CMC dressings did not kill bacteria, they immobilized them and prevented them from replicating, as there was no increase in numbers over 20 hours.15 Likewise, a study investigating the binding capacity of different strains of bacteria to dressings coated with DACC determined that microbes, including P. aeruginosa, once bound to the dressing, did not multiply.18
A limitation of the current study is that the study duration of 6 hours is relatively short compared with wound dressing use in a clinical setting, where they may remain in contact with the wound for several days. The duration of this study was, however, limited by the in vitro setting, as a longer duration would have resulted in further multiplication of the organisms on the filter or gauze, due to the lack of antimicrobial agents present in the dressings tested, making any standardization and hence interpretation of the results difficult. Another limitation is that the models only included single-species bacterial biofilm, whereas it is well known that wound biofilm is often polymicrobial and may also include yeasts, fungi and molds. However, single-species filter disc and gauze-biofilm models have been used in studies published previously, demonstrating that they are accepted as appropriate methods.25–27 These are all future study adaptations that could be performed; in addition, other biofilm models could be used, eg, Centers for Disease Control (CDC) reactor, colony biofilm drip flow reactor, or Lubbock chronic wound biofilm model.28,29
The ability of CMC dressings to most effectively remove antibiotic-resistant P. aeruginosa and MRSA bacteria in both planktonic and biofilm forms suggests they may play an effective role in reducing bioburden in the management of acute and hard-to-heal wounds in a clinical setting. Our study has tested a small but representative number of different dressing technologies, yet there are other dressings that claim to sequester or retain bacteria30 and hence further testing of such dressings is warranted.
Data Sharing Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Acknowledgments
The authors wish to thank Nicola Burke and Louise Jelliman for assisting with laboratory testing, Lorraine Ralph and Kenny Tran at Convatec for their medical writing assistance, and Phil Bowler for scoping the initial laboratory studies. This study was funded by Convatec.
Disclosure
All authors are employees at Convatec. KM, VLT and DGM own Convatec stock. The authors report no other conflicts of interest in this work.
References
1. Palmer J, Flint S, Brooks J. Bacterial cell attachment, the beginning of a biofilm. J Ind Microbiol Biotechnol. 2007;34(9):577–588. doi:10.1007/s10295-007-0234-4
2. Becker P, Hufnagle W, Peters G, Herrmann M. Detection of differential gene expression in biofilm-forming versus planktonic populations of Staphylococcus aureus using micro-representational-difference analysis. Appl Environ Microbiol. 2001;67(7):2958–2965. doi:10.1128/AEM.67.7.2958-2965.2001
3. Sauer K. The genomics and proteomics of biofilm formation. Genome Biol. 2003;4(6):219. doi:10.1186/gb-2003-4-6-219
4. Clinton A, Carter T. Chronic wound biofilms: pathogenesis and potential therapies. Lab Med. 2015;46(4):277–284. doi:10.1309/LMBNSWKUI4JPN7SO
5. Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol. 2004;2(2):95–108. doi:10.1038/nrmicro821
6. Probst S, Apelqvist J, Bjarnsholt T, Lipsky BA, Ousey K, P EJG. Antimicrobials and non-healing wounds: an update. J Wound Manage. 2022;23(S Sup 1):S1–S33. doi:10.35279/jowm2022.23.03.sup01
7. Hurlow J, Bowler PG. Acute and chronic wound infections: microbiological, immunological, clinical and therapeutic distinctions. J Wound Care. 2022;31(5):436–445. doi:10.12968/jowc.2022.31.5.436
8. Malone M, Bjarnsholt T, McBain AJ, et al. The prevalence of biofilms in chronic wounds: a systematic review and meta-analysis of published data. J Wound Care. 2017;26(1):20–25. doi:10.12968/jowc.2017.26.1.20
9. Schwarzer S, James GA, Goeres D, et al. The efficacy of topical agents used in wounds for managing chronic biofilm infections: a systematic review. J Infect. 2020;80(3):261–270. doi:10.1016/j.jinf.2019.12.017
10. Malone M, Johani K, Jensen SO, et al. Effect of cadexomer iodine on the microbial load and diversity of chronic non-healing diabetic foot ulcers complicated by biofilm in vivo. J Antimicrob Chemother. 2017;72(7):2093–2101. doi:10.1093/jac/dkx099
11. Fletcher J, Edwards-Jones V, Fumarola S, et al. Best practice statement: antimicrobial stewardship strategies for wound management. Wounds UK; 2020. Available from: https://www.wounds-uk.com/resources/details/best-practice-statement-antimicrobial-stewardship-strategies-wound-management.
12. Murphy CA, Atkin L, Swanson T, et al. Defying hard-to-heal wounds with an early antibiofilm intervention strategy: wound hygiene. J Wound Care. 2020;29(Suppl 3b):S1–S28. doi:10.12968/jowc.2020.29.Sup3b.S1
13. Bowler PG, Jones SA, Davies BJ, Coyle E. Infection control properties of some wound dressings. J Wound Care. 1999;8(10):499–502. doi:10.12968/jowc.1999.8.10.26356
14. Walker M, Hobot JA, Newman GR, Bowler PG. Scanning electron microscopic examination of bacterial immobilisation in a carboxymethyl cellulose (AQUACEL) and alginate dressings. Biomaterials. 2003;24(5):883–890. doi:10.1016/S0142-9612(02)00414-3
15. Newman GR, Walker M, Hobot JA, Bowler PG. Visualisation of bacterial sequestration and bactericidal activity within hydrating Hydrofiber wound dressings. Biomaterials. 2006;27(7):1129–1139. doi:10.1016/j.biomaterials.2005.07.046
16. Neilsen JT, Fogh K. Clinical utility of foam dressings in wound management: a review. Chronic Wound Care Manage Res. 2015;2:31–38. doi:10.2147/CWCMR.S50832
17. Ljungh A, Yanagisawa N, Wadstrom T. Using the principle of hydrophobic interaction to bind and remove wound bacteria. J Wound Care. 2006;15(4):175–180. doi:10.12968/jowc.2006.15.4.26901
18. Cooper R, Jenkins L. Binding of two bacterial biofilms to dialkyl carbamoyl chloride (DACC)-coated dressings in vitro. J Wound Care. 2016;25(2):76, 78–82. doi:10.12968/jowc.2016.25.2.76
19. Uzun M, Anand SC, Shah T. In vitro characterisation and evaluation of different types of wound dressing materials. J Biomed Eng Technol. 2023;1(1):1–7. doi:10.12691/jbet-1-1-1
20. United States Pharmacopeia. 61. Microbiological examination of nonsterile products: microbial enumeration tests. USP-NF; 2022.
21. Merritt JH, Kadouri DE, O’Toole GA. Growing and analyzing static biofilms. Curr Protoc Microbiol. 2005. doi:10.1002/9780471729259.mc01b01s00
22. Flemming HC, Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8(9):623–633. doi:10.1038/nrmicro2415
23. Ryder C, Byrd M, Wozniak DJ. Role of polysaccharides in Pseudomonas aeruginosa biofilm development. Curr Opin Microbiol. 2007;10(6):644–648. doi:10.1016/j.mib.2007.09.010
24. Hurlow J, Blanz E, Gaddy JA. Clinical investigation of biofilm in non-healing wounds by high resolution microscopy techniques. J Wound Care. 2016;25(Suppl 9):S11–S22. doi:10.12968/jowc.2016.25.Sup9.S11
25. Kalan LR, Pepin DM, Ul-Haq I, Miller SB, Hay ME, Precht RJ. Targeting biofilms of multidrug-resistant bacteria with silver oxynitrate. Int J Antimicrob Agents. 2017;49(6):719–726. doi:10.1016/j.ijantimicag.2017.01.019
26. Bowler PG, Parsons D. Combatting wound biofilm and recalcitrance with a novel anti-biofilm Hydrofiber wound dressing. Wound Med. 2016;14:6–11. doi:10.1016/j.wndm.2016.05.005
27. Parsons D, Meredith K, Rowlands VJ, Short D, Metcalf DG, Bowler PG. Enhanced performance and mode of action of a novel antibiofilm hydrofiber(R) wound dressing. Biomed Res Int. 2016;2016:7616471. doi:10.1155/2016/7616471
28. Goeres DM, Hamilton MA, Beck NA, et al. A method for growing a biofilm under low shear at the air-liquid interface using the drip flow biofilm reactor. Nat Protoc. 2009;4(5):783–788. doi:10.1038/nprot.2009.59
29. Sun Y, Dowd SE, Smith E, Rhoads DD, Wolcott RD. In vitro multispecies Lubbock chronic wound biofilm model. Wound Repair Regen. 2008;16(6):805–813. doi:10.1111/j.1524-475X.2008.00434.x
30. Singh G, Byrne C, Thomason H, McBain AJ. Investigating the microbial and metalloprotease sequestration properties of superabsorbent wound dressings. Sci Rep. 2022;12(1):4747. doi:10.1038/s41598-022-08361-3
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.