Back to Journals » Psoriasis: Targets and Therapy » Volume 13
The Skin Microbiome and Its Role in Psoriasis: A Review
Authors Celoria V, Rosset F , Pala V, Dapavo P, Ribero S, Quaglino P, Mastorino L
Received 11 July 2023
Accepted for publication 18 October 2023
Published 26 October 2023 Volume 2023:13 Pages 71—78
DOI https://doi.org/10.2147/PTT.S328439
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Uwe Wollina
Valentina Celoria,* Francois Rosset,* Valentina Pala, Paolo Dapavo, Simone Ribero, Pietro Quaglino, Luca Mastorino
Dermatologic Clinic, Department of Medical Sciences, University of Turin, Turin, Italy
*These authors contributed equally to this work
Correspondence: Valentina Celoria, Dermatologic Clinic, Department of Medical Sciences, University of Turin, Turin, Italy, Email [email protected]
Abstract: The skin microbiome is made of various microorganisms, most of which have the function of protecting individuals from harmful pathogens, and they are involved in innate and adaptive immune responses. The skin acts as a physical and immunological barrier against external stimuli, including pathogens and physical damage. Changes in the composition of the skin microbiome can trigger inflammatory processes leading to inflammatory skin diseases in susceptible individuals. Psoriasis (PsO) is a chronic inflammatory disease with a multifactorial etiology, where breakdown of immune tolerance to cutaneous microorganisms is implicated in its pathogenesis. Dysregulation of the microbiome due to genetic and environmental factors plays a significant role in the development of psoriatic disease. Dermatologic conditions such as atopic dermatitis, acne, psoriasis, and rosacea have been associated with intestinal dysbiosis. The skin microbiota composition is crucial for the development of appropriate immune responses, and alterations in the skin microbiome can contribute to changes in physiology and susceptibility to skin diseases or inflammatory conditions. Understanding the microbial settlement of the skin and the network of interactions that occur throughout life is essential for comprehending the pathogenesis of skin diseases and developing innovative treatments. With this article we tried to explore the relationship between the human microbiome and psoriatic disease, shedding light on the functions of the microbiome and the inflammatory disease processes to identify additional therapeutic targets. This review aims to highlight the relationship between skin and gut microbiome functions and inflammatory processes in skin psoriasis and psoriatic arthritis (PsA). The goal is to facilitate future studies on the skin microbiome to identify potential novel therapies for patients with psoriatic disease.
Keywords: psoriasis, skin microbiome, new therapies, gut microbiome, molecular precision medicines, next generation treatments
Background
The skin microbiome is composed of various microorganisms, most of which have the function of protecting individuals from harmful pathogens, and they are involved in innate and adaptive immune responses.1,2 The skin acts as a physical and immunological barrier against external stimuli, including pathogens and physical damage. Changes in the composition of the skin microbiome can trigger inflammatory processes in the epithelial microenvironment, leading to inflammatory skin diseases in susceptible individuals. Psoriasis, a chronic inflammatory skin disease, is caused by the interaction between multiple genetic and environmental risk factors. While the exact mechanisms are not yet clear, both the cutaneous and intestinal microbial populations contribute to the pathogenesis of psoriatic disease, as their alteration can lead to pro-inflammatory processes in the intestine, skin, and joints.3
Psoriasis (PsO) is a chronic inflammatory disease with a multifactorial etiology, where breakdown of immune tolerance to cutaneous microorganisms is implicated in its pathogenesis. Dysregulation of the microbiome due to genetic and environmental factors plays a significant role in the development of psoriatic disease. Dysbiosis, or an alteration in the composition of the microbiome, can lead to downstream proinflammatory effects in the gut, skin, and joints. Both the cutaneous and intestinal microbial populations are implicated in the pathogenesis of psoriatic disease, although the exact mechanisms are still unclear. This review aims to explore the relationship between the human microbiome and psoriatic disease, shedding light on the functions of the microbiome and the inflammatory disease processes to identify additional therapeutic targets. Furthermore, cutaneous dysbiosis has been observed not only in cutaneous psoriasis but also in Psoriatic arthritis (PsA).4
Materials and Methods
This review aims to highlight the relationship between skin and gut microbiome functions and inflammatory processes in skin psoriasis and psoriatic arthritis (PsA). The goal is to facilitate future studies on the skin microbiome to identify potential novel therapies for patients with psoriatic disease.5
The Skin Microbiome
In addition to the epithelial barrier, the gut microbiome also impacts the immune-regulatory properties of the gut. Certain intestinal microbes have the ability to produce or enhance the expression of immune-modulating molecules, including retinoic acid, polysaccharide A, and short-chain fatty acids (SCFAs), which play a role in maintaining the balance between effector and regulatory T cells. However, the specific microbes responsible for these mechanisms in PsO are not yet well understood.
The skin, the largest organ in the human body, serves as a barrier to prevent the invasion of pathogens. It harbors millions of bacteria, viruses, and fungi, collectively known as the skin microbiota, with bacterial density reaching cells per square centimeter. These bacteria play crucial roles in combating pathogens and contributing to the immune system.6,7 The colonization of the skin microbiome undergoes changes during the first year of life, particularly in the initial weeks after birth. This colonization process may impact long-term microbiome stability and skin function. For instance, the presence of the commensal bacterium Staphylococcus epidermidis during early life can reduce exposure to the pathogen S. aureus, thereby preventing inflammatory diseases.8 Bidirectional microbial transmission between mother and child is crucial in this process, while the role of paternal microbiota inheritance is yet to be elucidated. Moreover, the skin microbiome undergoes marked shifts during puberty, with a change in predominant bacterial groups and the overproduction of sebum associated with the overcolonization of Cutibacterium acnes.9 Over time, the skin microbiome undergoes age-related physiological changes, including alterations in sebum and sweat secretion, as well as changes in the immune response of the cutaneous barrier.10
When the skin barrier or the balance between commensal and pathogenic microorganisms is disrupted, various skin diseases or even systemic diseases can occur. Studying the microbiota composition at different skin sites is crucial to understanding the etiology of common skin disorders, which often manifest in specific areas of the skin.5 Advances in sequencing technology have enabled the analysis of the skin microbiota composition. Studies conducted on healthy adults have demonstrated associations between microbial communities and specific microenvironments. For example, lipophilic Propionibacterium species predominantly colonize sebaceous skin sites, while Staphylococcus and Corynebacterium are dominant in various areas, including the elbows and foot folds.11 Different skin sites exhibit considerable variation in bacterial diversity, with high diversity observed in the forearm, palm of the hand, index finger, back of the knee, and sole of the foot.11 Longitudinal studies have shown that the stability of the skin microbiome varies depending on the site, with less variability observed in the external auditory canal, inguinal crease, and wing crease, while more variability is seen in the popliteal fossa, forearm, and buttock.12 The microbial communities found on the hands are more similar to those on the forearm than those on the forehead or the inside of the elbow.13 Differences in the hand surface microbiome have been observed between men and women, and these differences become more evident as time since handwashing increases.13 Further studies are needed to elucidate the underlying factors contributing to these differences, including cutaneous pH, sweat or sebum production, use of creams, skin thickness, and hormonal variations.13
Temporal stability of the skin microbiome has been demonstrated in healthy individuals for up to 2 years, with the most abundant species contributing to the long-term stability of microbial communities, thus defining a unique microbial signature for each individual.14 Similar findings have been reported regarding the stability of the facial skin microbiome over a 2-year period, with substantial changes associated with alterations in stratum corneum barrier function and follicular porphyrins.15
Skin Microbiome and Skin Disorders
Using techniques described in seminal publications, researchers have gained new insights into the complexity of individual cutaneous microbiomes. Not only is the cutaneous microbiome highly diverse and individualized, but it is also tightly regulated within and between different skin regions, and stable over time. This stability has allowed for the identification of individual microbial signatures or “fingerprints” which have been the focus of research in forensic medicine, enabling the identification of individuals based on microbial information retrieved from surfaces they have touched.9
In literature, there are descriptions about the skin microbiome of injured and diseased skin, including psoriasis, and its substantial differences from healthy skin.9 Psoriasis is a multifactorial inflammatory disease with a complex pathogenesis, which in most cases manifests as well-circumscribed erythematous papules and plaques covered with silvery scales. The etiology of psoriasis is not entirely clear; it is hypothesized that an environmental trigger evokes a T-cell-mediated inflammatory response and subsequent hyper-proliferation of keratinocytes. Well-identified triggers include trauma in Koebner’s phenomenon, sunburns, HIV infection, Streptococcal infection in psoriasis guttata, medications such as beta-blockers or ACE inhibitors, emotional stresses, alcohol consumption, smoking, and obesity.16
Studies in the literature suggest that bacteria, including Staphylococcus aureus and Streptococcus pyogenes, can trigger and sustain psoriatic disease, and group A Streptococcus antigens and superantigens (GAS) have been implicated in the pathogenesis of psoriasis in individuals with genetic predisposition.17,18 In addition, the wounds on the skin caused by the severe itching of psoriatic lesions can promote the establishment of epidermal bacteria in the deep dermis and, in severe cases, into the bloodstream, where the encounter with immune system cells can promote an inflammatory environment and dysbiosis of the skin microbiota.19
These data, therefore, suggest the importance of the study of the skin microbiota of patients with psoriasis to improve the treatment of the disease.20
Gao and colleagues analyzed the microbiome of skin with psoriatic lesions, uninvolved skin, and skin from healthy people.21 The microbial population covering the lesions was more abundant and varied than the healthy skin samples, which were found to be relatively homogeneous. In addition, the lesions had a decrease in Actinobacteria, Proteobacteria, and Propionibacterium and its main human species P. acnes and an increase in Firmicutes, the three main phyla that populate healthy skin.22
Based on sequencing of the 16S rRNA V1-V3 variable region of 28 psoriasis patients and 26 healthy subjects, Chang et al showed that the relative abundance of the Staphylococcus species across all samples is associated with different disease states. They also confirmed that microbial communities in psoriatic lesions display higher heterogeneity but lower stability than healthy skin, associated with an increase of Proteobacteria, Pseudomonas genera, Staphylococcus aureus, and Staphylococcus pettenkoferi, and a reduction of Actinobacteria, Cutibacterium, Ethanoligenens, and Macrococcus genera, Cutibacterium acnes, Cutibacterium granulosum, and Staphylococcus epidermidis.23
On the other hand, Alekseyenko et al, comparing swabs of 54 patients with psoriasis and 37 healthy controls, demonstrated that the microbiome of psoriatic lesions is characterized by taxonomic diversity reduction and an increase of Firmicutes and Actinobacteria.24
Although there are several studies in the literature that have determined the composition of the microbiome of psoriatic skin compared with healthy skin, the results obtained are conflicting, particularly in relation to the variation in the abundances of Firmicutes, Actinobacteria, and Proteobacteria.25
Therefore, further studies are needed to better characterize the microbiome of psoriatic lesions and to establish standardized protocols that take into account hygiene regimen, climate, and cosmetic use, larger sample size, and standardize the amplification region and sampling method.26
Tobacco Smoke and Cutaneous Microbiome: Their Role in Psoriasis
Environmental factors are extremely important to consider in microbiome analysis. For example, smoking is known to promote the onset and maintenance of psoriasis and to reduce the response to treatment.27
In addition, patients with psoriasis have been known to be four times more likely to smoke. There are currently no specific studies in the literature on the effect of smoking on the skin microbiome and psoriasis.28 However, tobacco smoke is known to alter the lung and gastrointestinal microbiome, leading to a decrease in diversity in composition and favoring the presence of specific bacterial genera such as Bacteroides, Prevotella, Enterobacteria, and Clostridium. Since there is a potential interaction between the gastrointestinal and skin microbiomes, it is conceivable that tobacco smoke may directly and/or indirectly affect the skin microbiome, thereby contributing to the development of psoriasis. This possibility is supported by studies that have shown the presence of impaired gastrointestinal microbiomes in patients with inflammatory dermatoses.29
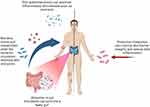
Skin and Gut Microbiome Crosstalk (Figure 1)
The skin and gut are complex immune and neuroendocrine organs that play a crucial role in maintaining the overall homeostasis and survival of the body.30 The composition of a healthy intestinal microbiome remains relatively stable from a young age and is characterized by the presence of Firmicutes, Bacteroidetes, Actinobacteria, Proteobacteria, Fusobacteria, and Verrucomicrobia, with Firmicutes and Bacteroidetes representing the majority. Gastrointestinal diseases and diet have been shown to have an impact on skin pathologies, as many dermatoses are strongly associated with gastrointestinal diseases.30 However, the precise mechanisms by which the gut microbiome affects skin health are still not fully understood.
Studies in mice have provided insights into the possible mechanisms involved in the relationship between the skin and gut microbiome. Administration of probiotics to mice has been shown to improve integumentary health, increase serum levels of IL-10 (an anti-inflammatory cytokine), and reduce IL-17 levels. Moreover, mice lacking IL-10 did not exhibit any modification of the integumentary system following probiotic administration, further supporting the involvement of IL-10 in the interaction between the skin and gut microbiome, possibly through the modulation of regulatory T cells (Tregs).30 Numerous studies in the literature support the hypothesis of skin-gut crosstalk mediated by the modulation of the immune environment through the microbiota. It has also been demonstrated that the intestinal microbiome can directly influence skin physiology and immune responses by the migration of microbiota and its metabolites to the skin.30
In recent years, there has been a significant increase in metagenomic data due to advances in sequencing technology, particularly in the field of skin microbiome research. The introduction of next-generation sequencing has revolutionized our understanding of the skin microbiome, allowing for a more comprehensive characterization of the diverse range of bacteria that colonize human skin 46. Specifically, the use of 16S rRNA sequencing has greatly expanded our understanding beyond what was previously possible with traditional culture techniques, which only identified about 1% of the cutaneous microbiome. Through 16S rRNA sequencing, a large international effort was able to chart the human microbiome by sampling diverse body habitats.
In cases where the intestinal barrier is compromised, the intestinal microbiota can enter the bloodstream and accumulate in the skin, leading to alterations. This hypothesis was supported by the detection of intestinal bacterial DNA in the plasma of patients with psoriasis.31
Literature studies have assessed the association between gut microbiome alterations and psoriasis. Chen et al observed an increase in Firmicutes and a decrease in Bacteroidetes in the intestines of psoriatic patients compared to control patients.32 Similar findings were reported by Huang et al, who found increased abundance of Firmicutes, Proteobacteria, and Actinobacteria, along with a decrease in Bacteroidetes in the gut microbiome of psoriatic patients.33 Scher et al evaluated gut microbiota alterations in patients with psoriatic arthritis and psoriasis and found a reduction in microbiota biodiversity and specific bacterial genera, such as Akkermansia, Ruminococcus, and Pseudobutyrivibrio, compared to healthy controls. The dysbiosis observed in these patients was associated with higher levels of secretory immunoglobulin A (sIgA) and a reduction in RANKL levels in the gut lumen of psoriatic arthritis patients.34 The authors speculated that the reduction of RANKL in PsA patients may be a response to the specific composition of the intestinal bacterial community.
Yu and colleagues investigated the causal relationship between gut microbiota and psoriasis and PsA using genome-wide association study (GWAS) data. Mendelian randomization analysis revealed that certain types of intestinal flora, including Lactococcus, Ruminiclostridium 5 and E. fissicatena are risk factors for psoriasis, while Odoribacter has shown a protective effect. The authors highlight a distinct set of risk and protective factors for PsA, despite it being a complication of psoriasis. In fact, Lactococcus, Verrucomicrobiales, Akkermansia, Coprococcus 1 and Verrucomicrobiaceae were found to be risk factors, while Odoribacter and Rikenellaceae have protective action against the development of PsA.35
Moreover, Zang and colleagues conducted two-sample Mendelian randomization study and observed that Bacteroidete and Prevotella 9 and Bacteroidia, Bacteroidales, and Ruminococcaceae UCG002 have protective action in psoriasis and PsA, respectively. The phylum Bacteroidetes is capable of producing short-chain fatty acids (SCFAs) that have anti-inflammatory activity and are reduced in psoriasis and PsA. On the other hand, Pasteurellales, Pasteurellaceae, Blautia, Methanobrevibacter, and Eubacterium fissicatena are risk factors for PsA, while E. fissicatena results as a risk factor for psoriasis after FDR correction.36
Probiotics and Effects on Skin
Several studies have demonstrated that probiotic intake can improve skin barrier integrity and reduce signs of reactive skin inflammation in mice.37 For instance, supplementation with Lactobacillus johnsonii protected glabrous mice from UV-induced contact hypersensitivity by reducing epidermal Langerhans cells and increasing systemic levels of IL-10.38 Similar protective effects were observed in a clinical trial with healthy volunteers after UV exposure, where intake of Lactobacillus johnsonii La1 normalized epidermal CD1A expression and maintained skin immune homeostasis.38
There are few clinical trials exploring the use of probiotics for the prevention and treatment of dermatological diseases, with the exception of atopic dermatitis. Most studies investigating probiotic interventions are oral, and of those that use topical probiotics, few include skin commensals. Generally, available clinical trials show positive results with improvements in skin conditions following probiotic intervention.
Oral and topical probiotics show promise in the treatment of specific inflammatory skin conditions and may have a beneficial effect on wound healing and skin cancer. Nevertheless, further research is required to verify these findings.
Although studies investigating the role of probiotics in psoriasis are lacking, there is some evidence that probiotics may have beneficial immunoregulatory effects by reducing inflammation. For example, in one study, oral administration of Bifidobacterium infantis for 8 weeks led to significantly decreased levels of inflammatory C-reactive protein and tumor necrosis factor-A in patients with psoriasis. However, it remains unclear whether this was accompanied by clinical improvements. In a mouse model of psoriasis, oral administration of Lactobacillus pentosus GMNL-77 reduced tumor necrosis factor-A and IL23-IL-17 axis cytokines, which was associated with decreased erythematous scaling lesions. Further research exploring the role of the microbiome and its modulation as a therapy in psoriasis would be a valuable complement to the many ongoing immunological treatment studies.
Fecal microbiota transplantation (FMT) has gained popularity in the past decade. Although FMT has mainly been used to treat gastrointestinal diseases, a few studies have explored its potential therapeutic benefits in psoriasis as well.39 One study reported significant improvement in severe plaque psoriasis and irritable bowel syndrome in a patient treated with FMT.40 Another study transplanted fecal microbiota from psoriatic patients and healthy individuals into mouse models of psoriasis and observed delayed recovery of psoriatic dermatitis and higher levels of IL-17A in mice receiving psoriatic microbiota.41 These findings suggest that FMT may hold promise as a therapeutic option for psoriasis, but further research is needed.42
Molecular studies in psoriatic patients have revealed biomarkers of an altered intestinal barrier, including higher levels of claudin-3 and intestinal fatty acid-binding protein.42 Integration of probiotics has shown improvements in psoriasis by increasing TNF-α expression, enhancing barrier function, and regulating the NF-kβ pathway involved in psoriasis pathogenesis.43,44
Conclusions
Dermatologic conditions such as atopic dermatitis, acne, psoriasis, and rosacea have been associated with intestinal dysbiosis. The skin microbiota composition is crucial for the development of appropriate immune responses, and alterations in the skin microbiome can contribute to changes in physiology and susceptibility to skin diseases or inflammatory conditions. Understanding the microbial settlement of the skin and the network of interactions that occur throughout life is essential for comprehending the pathogenesis of skin diseases and developing innovative treatments. Collecting data on the skin microbiome can aid in monitoring skin conditions, predicting disease onset, and selecting appropriate therapeutic approaches.
The role of the microbiota and its metabolic activity in psoriasis is gaining importance and may lead to the identification of crucial psoriatic biomarkers and the development of new therapeutic approaches. However, larger clinical studies with standardized methodologies are necessary to fully understand the role of the cutaneous microbiota and microbial pathways in psoriasis.
Probiotics have demonstrated an important role in promoting a healthy microbiome by reducing inflammation, modulating immune activation, and inhibiting the colonization of harmful bacteria. However, long-term safety data on probiotic use are limited, and caution is warranted, particularly in immunocompromised individuals who may be at risk of infections and serious side effects. Combination therapies involving phage or antibiotics may hold promise for microbiome replacement strategies and should be further explored.
In conclusion, there is a need for further research to fully understand the role of the microbiome in the development and treatment of diseases. Basic research and epidemiological studies are needed to identify the specific microbiota associated with disease and their mechanisms of action. Clinical trials with larger samples and greater power are necessary to establish the safety and efficacy of probiotic interventions, including identifying optimal species combinations, doses, and treatment durations. Additionally, exploring combination therapy with phages or antibiotics may provide promising results for microbiome replacement strategies. Overall, continued research in this field may lead to the development of new diagnostic and therapeutic opportunities for a range of diseases.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Belkaid Y, Segre JA. Dialogue between skin microbiota and immunity. Science. 2014;346(6212):954–959. doi:10.1126/science.1260144
2. Grice EA. The intersection of microbiome and host at the skin interface: genomic- and metagenomic-based insights. Genome Res. 2015;25(10):1514–1520. doi:10.1101/gr.191320.115
3. Chen L, Li J, Zhu W, et al. Skin and Gut Microbiome in Psoriasis: gaining Insight Into the Pathophysiology of It and Finding Novel Therapeutic Strategies. Front Microbiol. 2020;11:589726. doi:10.3389/fmicb.2020.589726
4. Zhang X, Shi L, Sun T, Guo K, Geng S. Dysbiosis of gut microbiota and its correlation with dysregulation of cytokines in psoriasis patients. BMC Microbiol. 2021;21(1). doi:10.1186/S12866-021-02125-1
5. Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326(5960):1694–1697. doi:10.1126/science.1177486
6. Grice EA, Segre JA. The skin microbiome. Nat Rev Microbiol. 2011;9(8):626. doi:10.1038/nrmicro2537.
7. Oh J, Byrd AL, Deming C, et al. Biogeography and individuality shape function in the human skin metagenome. Nature. 2014;514(7520):59–64. doi:10.1038/nature13786
8. Fierer N, Hamady M, Lauber CL, Knight R. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc Natl Acad Sci U S A. 2008;105(46):17994–17999. doi:10.1073/pnas.0807920105
9. Hillebrand GG, Dimitriu P, Malik K, et al. Temporal Variation of the Facial Skin Microbiome: a 2-Year Longitudinal Study in Healthy Adults. Plast Reconstr Surg. 2021;147(1S–2):50S–61S. doi:10.1097/PRS.0000000000007621
10. Luna PC. Skin Microbiome as Years Go By. Am J Clin Dermatol. 2020;21(Suppl 1):12–17. doi:10.1007/s40257-020-00549-5
11. Zhou Y, Mihindukulasuriya KA, Gao H, et al. Exploration of bacterial community classes in major human habitats. Genome Biol. 2014;15(5):R66. doi:10.1186/gb-2014-15-5-r66.
12. Grice EA, Kong HH, Conlan S, et al. Topographical and temporal diversity of the human skin microbiome. Science. 2009;324(5931):1190–1192. doi:10.1126/science.1171700
13. Dimitriu PA, Iker B, Malik K, Leung H, Mohn WW, Hillebrand GG. New Insights into the Intrinsic and Extrinsic Factors That Shape the Human Skin Microbiome. mBio. 2019;10(4):e00839–19. doi:10.1128/mBio.00839-19.
14. Edmonds-Wilson SL, Nurinova NI, Zapka CA, Fierer N, Wilson M. Review of human hand microbiome research. J Dermatol Sci. 2015;80(1):3–12. doi:10.1016/j.jdermsci.2015.07.006
15. Oh J, Byrd AL, Park M, Comparative Sequencing Program NISC, Kong HH, Segre JA. Temporal Stability of the Human Skin Microbiome. Cell. 2016;165(4):854–866. doi:10.1016/j.cell.2016.04.008
16. Vičić M, Kaštelan M, Brajac I, Sotošek V, Massari LP. Current Concepts of Psoriasis Immunopathogenesis. AMA Arch Dermatol. 2009;145(6):625–631.
17. Zhou X, Chen Y, Cui L, Shi Y, Guo C. Advances in the pathogenesis of psoriasis: from keratinocyte perspective. Int J Mol Sci. 2021;22(9):4615. doi:10.3390/ijms22094615
18. Gao Z, Tseng CH, Strober BE, et al. Substantial alterations of the cutaneous bacterial biota in psoriatic lesions. PLoS One. 2008;3(7):e2719. doi:10.1371/journal.pone.0002719
19. Tomi NS, Kranke B, Aberer E. Staphylococcal toxins in patients with psoriasis, atopic dermatitis, and erythroderma, and in healthy control subjects. J Am Acad Dermatol. 2005;53(1):67–72. doi:10.1016/j.jaad.2005.02.034
20. Lober CW, Belew PW, Rosenberg EW, Bale G. Patch tests with killed sonicated microflora in patients with psoriasis. Arch Dermatol. 1982;118(5):322–325. doi:10.1001/archderm.1982.01650170036019
21. Fahlen A, Engstrand L, Baker BS, Powles A, Fry L. Comparison of bacterial microbiota in skin biopsies from normal and psoriatic skin. Arch Dermatol Res. 2012;304(1):15–22. doi:10.1007/s00403-011-1189-x
22. Chang HW, Yan D, Singh R, et al. Alteration of the cutaneous microbiome in psoriasis and potential role in Th17 polarization. Microbiome. 2018;6(1):154. doi:10.1186/s40168-018-0533-1
23. Alekseyenko AV, Perez-Perez GI, De Souza A, et al. Community differentiation of the cutaneous microbiota in psoriasis. Microbiome. 2013;1(1):31. doi:10.1186/2049-2618-1-31
24. Drago L, De Grandi R, Altomare G, Pigatto P, Rossi O, Toscano M. Skin microbiota of first cousins affected by psoriasis and atopic dermatitis. Clin Mol Allergy. 2016;14:4. doi:10.1186/s12948-016-0038-z
25. Fyhrquist N, Muirhead G, Prast-Nielsen S, et al. Microbe-host interplay in atopic dermatitis and psoriasis. Nat Commun. 2019;10(1):4703. doi:10.1038/s41467-019-12253-y
26. Quan C, Chen XY, Li X, et al. Psoriatic lesions are characterized by higher bacterial load and imbalance between Cutibacterium and Corynebacterium. J Am Acad Dermatol. 2020;82(4):955–961. doi:10.1016/j.jaad.2019.06.024
27. Assarsson M, Soderman J, Dienus O, Seifert O. Significant differences in the bacterial microbiome of the pharynx and skin in patients with psoriasis compared with healthy controls. Acta Derm Venereol. 2020;100(8):adv00273. doi:10.2340/00015555-3619
28. Wei J, Zhu J, Xu H, et al. Alcohol consumption and smoking in relation to psoriasis: a Mendelian randomization study. J Invest Dermatol. 2021;141(4):984–991.e2.
29. Roth RR, James WD. Microbial ecology of the skin. Annu Rev Microbiol. 1988;42:441–464. doi:10.1146/annurev.mi.42.100188.002301
30. Lazaridou E, Giannopoulou C, Fotiadou C, Vakirlis E, Trigoni A, Ioannides D. The potential role of microorganisms in the development of rosacea. J Dtsch Dermatol Ges. 2011;9(1):21–25. doi:10.1111/J.1610-0387.2010.07513.X
31. Chen YJ, Wu CY, Chen JD, et al. The risk of cancer in patients with psoriasis: a population-based cohort study in Taiwan. J Am Acad Dermatol. 2011;64(2):276–282.
32. Huang C, Yang YJ, Chen YY, et al. Soluble epoxide hydrolase activity and pharmacologic inhibition in horses with chronic severe laminitis. Am J Vet Res. 2011;72(3):412–421.
33. Scher JU, Ubeda C, Artacho A, et al. Decreased bacterial diversity characterizes the altered gut microbiota in patients with psoriatic arthritis, resembling dysbiosis in inflammatory bowel disease. Arthritis Rheumatol. 2015;67(1):128–139. doi:10.1002/art.38892
34. Yu Y, Dunaway S, Champer J, Kim J, Alikhan A. Changing our microbiome: probiotics in dermatology. Br J Dermatol. 2020;182(1):39–46. doi:10.1111/BJD.18088
35. Nianzhou Y, Wang J, Liu Y, Guo Y. Investigating the gut microbiota’s influence on psoriasis and psoriatic arthritis risk: a Mendelian randomization analysis. Precision Clin Med. 2023;6(3):pbad023. doi:10.1093/pcmedi/pbad023
36. Zang C, Liu J, Mao M, Zhu W, Chen W, Wei B. Causal Associations Between Gut Microbiota and Psoriasis: a Mendelian Randomization Study. Dermatol Ther (Heidelb). 2023;13(10):2331–2343. doi:10.1007/s13555-023-01007-w
37. Guéniche A, Benyacoub J, Buetler TM, Smola H, Blum S. Supplementation with oral probiotic bacteria maintains cutaneous immune homeostasis after UV exposure. Eur J Dermatol. 2006;16(5):511–517.
38. Raychaudhuri SK, Maverakis E, Raychaudhuri SP. Diagnosis and classification of psoriasis. Autoimmun Rev. 2014;13(4–5):490–495. doi:10.1016/J.AUTREV.2014.01.008
39. Wang JW, Kuo CH, Kuo FC, et al. Fecal microbiota transplantation: review and update. J Formos Med Assoc. 2019;118(Suppl 1):S23–S31. doi:10.1016/J.JFMA.2018.08.011
40. Dougan SK, Kaser A, Blumberg RS. CD1 expression on antigen-presenting cells, in T Cell Activation by CD1 and Lipid Antigens. In: Current Topics in Microbiology and Immunology. Vol. 314. Berlin: Springer; 2007.
41. Peguet-Navarro J, Dezutter-Dambuyant C, Buetler TM, et al. Supplementation with oral probiotic bacteria protects human cutaneous immune homeostasis after UV exposure - double-blind, randomized, placebo controlled clinical trial. Eur J Dermatol. 2008;18(5):504–511. doi:10.1684/ejd.2008.0496
42. Polkowska-Pruszyńska B, Gerkowicz A, Krasowska D. The gut microbiome alterations in allergic and inflammatory skin diseases - an update. J Eur Acad Dermatol Venereol. 2019;34(3):455–464. doi:10.1111/jdv.15951
43. Nayak RR. Western Diet and Psoriatic-Like Skin and Joint Diseases: a Potential Role for the Gut Microbiota. J Invest Dermatol. 2021;141(7):1630–1632. doi:10.1016/J.JID.2021.01.003
44. Widhiati S, Purnomosari D, Wibawa T, Soebono H. The role of gut microbiome in inflammatory skin disorders: a systematic review. Dermatol Rep. 2021;14(1). doi:10.4081/DR.2022.9188
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.