Back to Journals » Nanotechnology, Science and Applications » Volume 7
Prevention of bacterial foodborne disease using nanobiotechnology
Authors Billington C, Hudson A, D'Sa E
Received 21 March 2014
Accepted for publication 27 May 2014
Published 25 August 2014 Volume 2014:7 Pages 73—83
DOI https://doi.org/10.2147/NSA.S51101
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Craig Billington, J Andrew Hudson, Elaine D'Sa
Food Safety Programme, ESR, Ilam, Christchurch, New Zealand
Abstract: Foodborne disease is an important source of expense, morbidity, and mortality for society. Detection and control constitute significant components of the overall management of foodborne bacterial pathogens, and this review focuses on the use of nanosized biological entities and molecules to achieve these goals. There is an emphasis on the use of organisms called bacteriophages (phages: viruses that infect bacteria), which are increasingly being used in pathogen detection and biocontrol applications. Detection of pathogens in foods by conventional techniques is time-consuming and expensive, although it can also be sensitive and accurate. Nanobiotechnology is being used to decrease detection times and cost through the development of biosensors, exploiting specific cell-recognition properties of antibodies and phage proteins. Although sensitivity per test can be excellent (eg, the detection of one cell), the very small volumes tested mean that sensitivity per sample is less compelling. An ideal detection method needs to be inexpensive, sensitive, and accurate, but no approach yet achieves all three. For nanobiotechnology to displace existing methods (culture-based, antibody-based rapid methods, or those that detect amplified nucleic acid) it will need to focus on improving sensitivity. Although manufactured nonbiological nanoparticles have been used to kill bacterial cells, nanosized organisms called phages are increasingly finding favor in food safety applications. Phages are amenable to protein and nucleic acid labeling, and can be very specific, and the typical large "burst size" resulting from phage amplification can be harnessed to produce a rapid increase in signal to facilitate detection. There are now several commercially available phages for pathogen control, and many reports in the literature demonstrate efficacy against a number of foodborne pathogens on diverse foods. As a method for control of pathogens, nanobiotechnology is therefore flourishing.
Keywords: bacteriophage, pathogen control, detection, food safety
Introduction
The impact of foodborne disease is a significant economic and clinical issue, despite recent advances in food safety. In clinical terms, the number of cases per year in the US has been estimated at 9.4 million, with 55,961 hospitalizations and 1,351 deaths.1 The annual consequent cost of foodborne illness is $51 billion to $77.7 billion, depending on the model used. Reducing the occurrence of foodborne illness through the use of rapid, cost-effective detection procedures and new ways to control pathogens is the focus of much current research.
Nanobiotechnology can be defined as the intersection of nanotechnology and biotechnology. This concise review article considers nanobiotechnological detection of, and interventions to control, bacterial foodborne pathogens. We have chosen to focus on nanostructures that either incorporate biological elements or are entirely biological in origin, and particularly on applications of bacteriophages (phages).2 We have excluded nonbiological materials such as silver nanoparticles that are used to kill pathogens and have been reviewed elsewhere.3 We also acknowledge that many examples and applications have been omitted from this review because of space limitations, and readers are referred to the many excellent recent reviews that are available.
Detection of bacterial pathogens
In an ideal world it would be desirable to detect even a single pathogen present on/in a food sample. Current “traditional” culture-based methods tend to use an enrichment step in which low numbers of target cells are encouraged to multiply to detectable numbers. Enrichment also ensures that any bacteria detected are alive. This approach allows a theoretical detection limit of 0.04 cell/g (ie, one cell in 25 g, a normal analytical weight). However, enrichment typically adds 1–2 days to the total analysis time. When enrichment is not used, it is usual for a food sample to be diluted one in ten prior to analysis, thus also reducing the cell concentration available for detection tenfold (eg, ten cells/g of food become one cell/mL in the diluent).
It is imperative to have methods that are able to detect low concentrations of pathogens, as it can sometimes take only a few cells in a food to cause disease (Table 1). Another factor to be considered is the accuracy of testing methods, as judged by the proportion of false positive and false negative results. For a method to be adopted widely it is usual for it to receive official validation. An example of this is a “performance-tested method” by AOAC Research Institute (http://goo.gl/413U6y). Receiving such validation endorses all the performance parameters of the test.
For nanobiotechnology to offer improvements over traditional methods it will need to allow the detection of pathogens faster, less expensively, and/or more accurately than existing methods. Ideally, it should be sufficiently sensitive to eliminate the need for enrichment, though probably requiring enhanced sample preparation methods. This need has been addressed in some papers. For example, pathogens have been detected at 20–50 colony-forming units (CFU)/mL within 2 hours without enrichment10 by prior concentration of bacteria in food rinses using immunomagnetic separation (IMS).
Assessment of methods cited in literature
Pathogen detection methods using nanobiotechnology that are cited in the literature need to be examined from the perspective of what they bring to a crowded rapid methods diagnostics market. For example, one study reports the detection of three pathogens at 400–800 cells/mL.11 For the suggested application in testing milk, this assay is too insensitive to be used without enrichment, so the associated improved test speed is largely negated.
Currently, there are rapid pathogen detection kits available that use antibodies to capture pathogens, and many of these produce a qualitative visual signal (eg, lateral flow devices) or a colored enzymatic reaction-derived product that can be measured by spectrophotometry (eg, enzyme-linked immunosorbent assay). In general, low-sensitivity methods are applicable only to detecting pathogens in enrichments. There are also many methods available that are based on the detection of amplified segments of nucleic acid (polymerase chain reaction [PCR]), which can be sensitive, rapid, and specific.
Quantum dots, nanobeads, and nanorods
There has been considerable research into the use of nanosized quantum dots (QDs) to detect foodborne pathogens. These semiconductor nanocrystals have been used as fluorophores for cellular imaging, as they possess superior properties to conventional fluorophores. QDs have been coupled with specific antibodies to facilitate detection of organisms, including the parasites Cryptosporidium parvum and Giardia lamblia12 and the bacteria Mycobacterium bovis,13 Escherichia coli O157:H7,14,15 Listeria monocytogenes,10 Salmonella,16 and Shigella.16 Indeed, a modified cellphone has been used as a detection system for E. coli.17 Toxins, including shiga-like toxin, cholera toxin, and ricin, have been detected using a QD protocol.18
The limit of detection of these QD methods is reported to be ~103–104 organisms/mL,13 depending on the detection method used, although a limit of five to ten organisms/mL has also been claimed.17 If an enrichment is used, the detection limit can be reduced to ten organisms/mL,14 but this also increases the test time.
Carboxylated magnetic nanobeads (diameter 180 nm) coated with antibodies have been used to detect Salmonella typhimurium, E. coli O157:H7, and L. monocytogenes in foods by multiplex PCR, but the detection limit, approaching 104 CFU/mL, means that enrichment of the food would be needed for practical application.19 Silicon nanorods onto which antibodies were attached have also been used to detect Salmonella,20 although fluorescence microscopy, a low throughput detection method, was used to detect the labeled cells. The limit of detection was not reported.
Antibody- and nucleic acid-based biosensors
Biosensors make use of antibodies or phage components (see Phage-based nanobiotechnological detection methods) that have specific recognition elements for pathogenic microbes. When the microbe is captured by the recognition element, a measurable response occurs. In these systems the antibody is attached to a suitable matrix in close proximity to a transducer, which can be optical, electrochemical, piezoelectric, or thermometric21 and which produces a measurable signal. Although biosensors may offer the prospect of near to real-time detection, they currently lack the sensitivity to be used without an initial enrichment (see Narsaiah et al),21 although they could be applied to some pathogens, such as Staphylococcus aureus, for which there is tolerance of moderate concentrations in some foods. As a specific example, a detection system for L. monocytogenes could detect a single cell,22 but the sample volume was only 4 μL, giving a theoretical detection limit in the food of 500 CFU/g, given the 50:50 sample dilution used. Although these systems can offer reuse of the biosensor,23 unlike lateral flow devices, the equipment requires significant capital outlay.
Biosensors may also use nucleic acid hybridization as a means of detecting bacteria. A recently described detection system for S. typhimurium used both carbon nanotubes and nanoparticles as components in an assay, detecting down to 1.5×102 CFU/mL of the pathogen in milk.24
The use of antibodies as a recognition/capture element of bacteria has been quite well described, so here we focus on the application of phage subcomponents for the same purpose.
Phage-based nanobiotechnological detection methods
Phages are naturally occurring nanosized killers of bacteria.25 The smallest group are icosohedra (diameter 27 nm), but they can be up to 2,400 nm long in other groups.26 Phages are environmentally common and are the most numerous biological entities on the planet.27 Their high specificity and lethal effect, and the relative ease of engineering their genomes and structures lend them to nanobiotechnological applications for food safety.
Rapid detection techniques have been developed using the bactericidal and specificity properties of free phage particles. Impedance–conductance methods have been combined with phages for the rapid detection of E. coli O157:H728 and Salmonella.29 Selective enrichments are performed and electrical properties of the media monitored for changes resulting from metabolism of substrates by target bacteria. The added phages retard the growth of the target bacterium, resulting in a characteristic change in electrical profile of the medium, so enabling rapid identification. These methods may be prone to false negative results and require careful tuning of conditions.
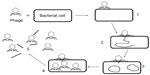
In a novel alternative technique,30 the sample containing target bacteria was first inoculated with a low concentration of phages for a short time (2 hours) to enable at least one phage replication cycle (Figure 1). Bacterial “helper cells” known to be susceptible to the phage were then added to the sample to amplify phage numbers during another short 1-hour to 2-hour incubation. The ratio of intact to damaged helper cells was measured by Live/Dead staining, enabling the calculation of the original number of cells present in the sample. The authors were able to detect 100 CFU/mL of Pseudomonas aeruginosa using this method.
The detection of progeny phages following infection of bacteria during enrichment is also an approach for rapid bacterial detection. Such assays have been used in vitro for the detection of Pseudomonas and Salmonella, with sensitivities of 40 CFU/mL and 600 CFU/mL, respectively, in 4 hours.31 A commercial assay for the detection of Mycobacterium avium subspecies paratuberculosis in milk is under development using this technique and is currently able to determine presence/absence within 48 hours32 (which is rapid for this slow-growing microorganism). Amplification assays have also been combined with mass spectrometry where phage proteins are the target for bacterial identification and detection. This approach has been demonstrated for the detection of E. coli and Salmonella,33 and for S. aureus,34 though improvements in the sensitivity would be required for commercial use.
A number of methods have been proposed for the detection of bacteria by phages that involve modification of the phage DNA or capsid (head) protein. The phage genome is relatively amenable to genetic engineering, and this has allowed the insertion of a number of reporter genes, facilitating rapid detection of bacteria. In these systems (summarized in Table 2), the engineered gene is expressed soon after bacterial infection, which enables both specific and rapid detection compared with conventional techniques. The sensitivity of these methods is good (eg, 1 CFU/mL with 6 hours’ enrichment),35 but a potential limitation is the need for phages with the correct specificity. Rather than detecting the expression of a gene in the target bacterium, an alternative is direct fluorescent DNA staining or phage capsid labeling. These techniques are flexible, in that the fluorescent signal can be detected by flow cytometry, multiwell plate readers, or direct microscopy. Goodridge et al45 described the use of IMS combined with fluorescent staining of a phage to detect 2.2 CFU/g E. coli O157:H7 in ground beef following a 6-hour enrichment and 10–100 CFU/mL E. coli O157:H7 in raw milk after 10 hours’ enrichment. The capsid of E. coli phage T7 has been labeled with QDs by conjugation with biotin,46 and the approach was able to detect as few as 20 CFU/mL E. coli in a water sample within 1 hour, compared with 24 hours with conventional techniques.
  | Table 2 Reporter genes used in phage detection of foodborne pathogens |
Immobilized phage-based devices
The immobilization of phages onto surfaces for pathogen detection has several advantages over using free particles, particularly by allowing the possibility of incorporating phages into high-throughput or hand-held devices. Phages have been used in a magnetoelastic biosensor to detect Salmonella on the surfaces of watermelons,47 tomatoes,48 and egg shells49 and in milk.50 The phages are coated onto the biosensor chip, and when Salmonella cells bind to the phages the mass of the biosensor increases. The change in mass and resultant decrease in resonance were measured when a surface-scanning coil detector was passed within close proximity. This technique raises the possibility of immobilizing sensors in food packaging that could then be checked for the presence of a pathogen by passing a detector across the product. The current limitation is the relative lack of sensitivity (limit of detection 150 CFU/mm2) compared with conventional methods.
Surface plasmon resonance (SPR) is a method that has been harnessed to detect pathogens using gold immobilized phages. SPR is an optical technique that involves measuring small changes in the refractive index at the boundary of two media when illuminated by polarized light. Like magnetoelastic sensors, this technique promises rapid, label-free detection of pathogens, but more work is required to improve sensitivity. Phage-based SPR biosensors have been used in vitro for the detection of E. coli K12,51 E. coli O157:H7,52 and S. aureus,52,53 with detection limits of ~1,000 CFU/mL in 20 minutes.
Laube et al54 described a phage-based detection method for Salmonella where a phage was coated onto paramagnetic beads. The beads were used to capture and concentrate Salmonella from milk samples, with or without enrichment, and the bacteria were detected using peroxidase conjugated to Salmonella-specific antibodies. Limits of detection were 19 CFU/mL with no enrichment (assay time 2.5 hours) and 1.4 CFU/mL with brief enrichment (assay time 6 hours), which are comparable with existing methods taking 24 hours.
One limitation of immobilized phage biosensors is the potential for false negative results arising from the failure to detect injured or viable but not culturable cells. Fernandes et al55 offer a possible solution to this with the use of a lytic phage bound to a magnetoresistive biochip and antibody-coated nanoparticles for detection. Lytic phages are usually not favored for biosensors, as they destroy the cell before it can be detected, but here the authors used physiological conditions enabling cells to survive long enough to be detected. They were able to detect injured cells using the biosensor.
One of the main challenges in the development of nanobiosensors using intact phage particles is to immobilize them at a sufficiently high density. An approach to circumvent this is to use phage-encoded proteins that have high affinity for the bacterial cell wall, either receptor proteins from phage tail fibers or the cell-binding domains (CBDs) from endolysins (for Gram-positive bacteria only). The receptor-binding protein from a Campylobacter phage has been immobilized and used with SPR to detect as few as 100 CFU/mL.56 Technology based on this principle has been developed by Hyglos GmbH (Bernried am Starnberger See, Germany) and licensed to bioMérieux (Craponne, France) for use in its VIDAS® detection systems for E. coli O157:H7, Salmonella, and Listeria.
Walcher et al57 immobilized the CBD from a Listeria phage endolysin onto paramagnetic beads and combined this with real-time PCR to develop a rapid method for detecting Listeria. They were able to recover 10–100 CFU/mL Listeria from raw milk spiked with each of five isolates tested. The same group also successfully immobilized a Listeria endolysin CBD on a gold screen-printed electrode for detection by electrochemical impedance spectroscopy, but sensitivity is currently ten- to 100-fold poorer.58
Control in food production and distribution
Phage-based control
Intuitively, it is clear that testing of food alone cannot be relied upon to assure food safety, and there is a need to inactivate (control) any pathogens that are present before distribution to the consumer. Phage-based pathogen control offers an attractive proposition for control purposes, as phages are naturally present in food, and there is accumulating evidence that they are both safe and effective.
For meat it would be optimal to intervene at the preharvest stage, as bacteria are spread to carcasses via direct or indirect cross-contamination from feces on the hide in ruminant animal processing,59 from feathers during mechanical plucking of poultry,60 and from the digestive tract of seafood during processing.61 Supporting this premise, modeling of preslaughter intervention strategies, such as the use of phages to reduce shedding of E. coli O157:H7 in cattle feces, had the largest positive impact on carcass contamination.62
Preharvest interventions using phage biocontrol have been trialed for a number of food animals, the most notable being applications for poultry. Bardina et al63 were able to reduce concentrations of Salmonella in chicken ceca by 4.4 log10 CFU/g in 2 days and up to 2 log10 CFU/g, compared with controls, at 8 days (end of experiment) following oral administration of phages. In a similar trial, Wong et al64 achieved a 2.9 log10 CFU/mL reduction in bacterial concentrations after 6 hours, with Salmonella becoming undetectable in the cecum at 24 hours. A novel approach taken by Waseh et al65 was to purify the receptor (tail-spike) protein from a phage and feed it to chickens, attaining a 1–2 log10 CFU/mL reduction in Salmonella concentration in chicken ceca.
Campylobacter is a significant food safety issue for the poultry industry,66 and there have been several promising recent trials using phages to control Campylobacter in chickens. A study performed in three commercial broiler houses by Kittler et al67 showed that one of three experimental groups exhibited a significant reduction in Campylobacter concentration of >3.2 log10 CFU/g cecal contents at slaughter. However, there was no difference in the other two groups. Carvalho et al68 achieved a 2 log10 CFU/g reduction in fecal Campylobacter concentration using oral gavage to deliver phages, but this was increased by 0.5 log10 when the phages were delivered in feed.
Shiga toxigenic E. coli O157:H7 is a potentially fatal foodborne pathogen, and its primary reservoirs are cattle and other ruminants.69 Phage-based preharvest interventions for ruminants have been studied extensively with mixed results, and several studies report small or insignificant effects. Nevertheless, a single oral administration of phages reduced E. coli O157:H7 in the gastrointestinal tract of sheep by up to 4 log10,70 and a multidose phage protocol reduced fecal shedding of E. coli O157:H7 in cattle by 1 log10 CFU/g for up to 72 days.71
OmniLytics, Inc. (Sandy, UT, USA) (http://www.omnilytics.com) has taken a different approach and produced a phage-based hide wash for cattle, Finalyse™ (marketed by Elanco, Greenfield, IN, USA), to control E. coli O157:H7. It appears to be widely used in the US, although independent verification of its efficacy is lacking. A study of the application of phages to cattle hides72 demonstrated that up to 1.5 log10 CFU/cm2 reduction in E. coli O157:H7 could be achieved on detached hides 1 hour post-treatment. Clearly, there appears to be promise in the use of phages for hide decontamination, but techniques to control the pathogen within the gastrointestinal tract will require some further development.
For economic and public health reasons, control of Vibrio spp. is important in aquaculture. Phage biocontrol of V. vulnificus in live shucked oysters achieved up to 5 log10 CFU reductions in pooled tissue using a nine-phage cocktail.73 Subsequently, the same cocktail was evaluated in vitro for the control of V. vulnificus either alone or with oyster extract.74 Over 24 hours, the phage cocktail reduced the concentration of V. vulnificus from 6 log10 to 3 CFU/mL at 4°C, and the addition of oyster extract had a small additive effect. Phages may therefore have potential as a biodepuration treatment for V. vulnificus in oysters and other shellfish.
The easiest and most common application of phages for food safety purposes is to add them directly to foods during processing, and some phage products are now available commercially. Listex™ (P100), produced and marketed by Micreos BV (Wageningen, the Netherlands) (http://www.micreos.com), targets L. monocytogenes, and its efficacy has been demonstrated for several foods (Table 3). Intralytix Inc. (Baltimore, MD, USA) (http://www.intralytix.com) markets a range of similar phage products, including ListShield™, EcoShield™ (formerly ECP-100), and SalmoFresh™.
  | Table 3 Examples of phage biocontrol of foodborne pathogens |
Beyond commercial products, phages have proven to be effective for pathogen biocontrol in a wide range of foods, including meat, dairy, fruit, and produce, and against most of the significant foodborne pathogens (Table 3). Other ways of harnessing phages to control pathogens include using inactivated particles, immobilizing particles on packaging, or immobilizing phage-encoded lytic proteins (endolysins). Hudson et al100 produced inactive (nonreplicating) phage particles that retained the ability to kill E. coli on meat. The use of inactive phage particles may have advantages for perceived safety or regulatory classification purposes.
Phages have been successfully immobilized onto a cellulose substrate101 and could control Listeria and E. coli O157:H7 on raw and ready-to-eat meat under different storage temperatures and food packaging conditions. A reduction in Listeria concentration of up to 4 log10 CFU/g and in E. coli O157:H7 of up to 1.2 log10 CFU/g was achieved. It has been reported that a variety of phages can be immobilized on silica particles102 and incorporated into electrospun fibres,103 both of which are potential applications in food packaging. Solanki et al104 immobilized phage endolysins on both silica and starch nanoparticles, and reductions of 4.0 log10 CFU/g and 1.2 log10 CFU/g Listeria, respectively, were recorded on iceberg lettuce. These nanoparticles may also be able to be incorporated onto the surfaces of equipment and into the infrastructure of food processing premises, in order to minimize cross-contamination.
Nonphage-based nanobiotechnological controls
Nanotechnology has begun to be explored as a means of targeted delivery of antimicrobial agents into food or as a means of protecting those agents from adverse environmental conditions prior to their application.105 A wide range of encapsulation delivery systems exists,106 but new developments in nanotechnology promise improvements over those that are currently available. Targeted delivery of antibacterial materials can be achieved by several means, including liposomes,107 archaeosomes, nanochelates, casein micelles, globular food proteins that can reassemble into nanofibrils at elevated temperatures and low pH values, and polymers, natural or synthetic, including nanospheres, nanocapsules,108 polymersomes, and micelles.109
One example of a biological material used in nanotechnology is chitosan, which is nontoxic and biodegradable. It is used because of its inherent antimicrobial nature,110–112 which increases at low pH (with increased protonation).113 In addition, chitosan nanoparticles (CNPs) can be used to deliver other biologically derived antibacterial agents such as carvacrol, a component of some essential oils, to produce a synergistic antimicrobial effect. CNPs allow for a burst of release of the antimicrobial compound (eg, oregano essential oil), followed by a sustained release over time.114
The antibacterial effects of oleoyl–chitosan nanoparticles (OCNPs) against E. coli and S. aureus have been explored.115 Variations in molecular weight and degree of substitution of the chitosan were shown to inhibit the two organisms differently, but inhibition was optimal at pH 6. Antibacterial activity against the same two organisms has also been reported for nisin-loaded CNPs.116 In this case, peak nisin release occurred at day 1, but then a residual concentration was maintained for up to 100 days, and it was claimed that the combination of nisin and the inherent antibacterial effect of chitosan was synergistic when compared with the effect of either alone. However, insufficient data were provided to rule out the possibility that the effect was additive rather than synergistic. The activity of chitosan,117 OCNPs118 and nisin loaded CNPs116 involves the disruption of cell membranes.
Encapsulation of more than one bioactive component within a nonmaterial to allow simultaneous targeted and controlled release of both has been researched. For example, the incorporation of organic acid and essential oil in a chitosan matrix inhibited the growth of bacteria, including Enterobacteriaceae, on vacuum-packed cured meat products.113
Chitosan can be used as a packaging film, but it has a number of drawbacks, including poor water resistance, gas barrier, and mechanical properties. However, when used in combination with nanoclay to form a bionanocomposite film, the properties of the resulting structure are improved. The addition of rosemary essential oil into this film resulted in antimicrobial activity against L. monocytogenes and spoilage organisms.119
There is increasing interest in the use of biologically based polymers for food packaging purposes,120 with nanoscale fillers, including layered silicate nanoclays, being developed. Such nanocomposite films can contain inorganic material such as silver nanoparticles or biologically derived antimicrobials like essential oils, enzymes, and bacteriocins.
It may also be possible to induce antibody responses in the food animal against pathogenic bacteria. This has been achieved in black seabream by the oral administration of a DNA nanoparticle vaccine to induce an immune response against Vibrio parahaemolyticus.121
Conclusion
The biggest challenge for the detection of pathogens is for new faster, more cost-effective methods to meet the sensitivity of conventional approaches. One way to achieve this might be to concentrate bacteria from food samples, ideally removing the need to use lengthy enrichment procedures. There may be aspects of nanobiotechnology that can help: eg, nanosized beads for IMS, which could be used to recover pathogens from complex samples prior to detection. Applications to detect bacteria that have some allowable level in foods, such as S. aureus, Bacillus spp., E. coli, and some clostridia, could be achieved more readily. Another challenge is to ensure that dead bacteria are not detected while injured bacteria are.
The use of advanced nanocomposite packaging films may help to combat foodborne disease if they can alert producers and consumers to the presence of pathogens or to deliver a sustained and targeted antimicrobial effect. As a control agent, phages show promise, as reflected in the appearance of commercially available products and a growing literature base. Drivers for adoption of bionanotechnologies for food safety include an increased awareness of the need for effective pathogen control by “green” technologies that do not negatively impact product quality.
Beyond food safety, applications in these technologies in human health may provide useful options in the future: eg, in the treatment of human infections caused by antibiotic-resistant bacteria.
Acknowledgments
We would like to thank Stephen On, Susan Paulin, and Jeremie Langlet for constructive criticism of the manuscript.
Disclosure
The authors report no conflicts of interest in this work.
References
Scallan E, Hoekstra RM, Angulo FJ, et al. Foodborne illness acquired in the United States: major pathogens. Emerg Infect Dis. 2011;17(1):7–15. | |
Sekhon BS. Nanotechnology in agri-food production. Nanotechnol Sci Appl. 2014;7:31–53. | |
Duncan TV. Applications of nanotechnology in food packaging and food safety: barrier materials, antimicrobials and sensors. J Colloid Interface Sci. 2011;363(1):1–24. | |
Hara-Kudo Y, Takatori K. Contamination level and ingestion dose of foodborne pathogens associated with infections. Epidemiol Infect. 2011;139(10):1505–1510. | |
Yde M, Naranjo M, Mattheus W, et al. Usefulness of the European epidemic intelligence information system in the management of an outbreak of listeriosis in Belgium, 2011. Euro Surveill. 2012;17(38).pii:20279. | |
Graves LM, Hunter SB, Ong AR, et al. Microbiological aspects of the investigation that traced the 1998 outbreak of listeriosis in the United States to contaminated hot dogs and establishment of molecular subtyping-based surveillance for Listeria monocytogenes in the PulseNet network. J Clin Microbiol. 2005;43(5):2350–2355. | |
Lehmacher A, Bockemühl J, Aleksic S. Nationwide outbreak of human salmonellosis in Germany due to contaminated paprika-powdered potato chips. Epidemiol Infect. 1995;115:501–511. | |
McCallum L, Paine S, Sexton K, et al. An outbreak of Salmonella typhimurium phage type 42 associated with the consumption of raw flour. Foodborne Pathog Dis. 2013;10(2):1–6. | |
Unicombe LE, Simmons G, Merritt TD, et al. Sesame seed products contaminated with Salmonella: three outbreaks associated with tahini. Epidemiol Infect. 2005;133:1065–1072. | |
Wang H, Li Y, Wang A, Slavik M. Rapid, sensitive and simultaneous detection of three foodborne pathogens using magnetic nanobead-based immunoseparation and quantum dot-based multiplex immunoassay. J Food Prot. 2011;74(11):2039–2047. | |
Viswanathan S, Rani C, Ho JA. Electrochemical immunosensor for multiplexed detection of food-borne pathogens using nanocrystal bioconjugates and MWCNT screen-printed electrode. Talanta. 2012;94(0):315–319. | |
Zhu L, Ang S, Liu W-T. Quantum dots as a novel immunofluorescent detection system for Cryptosporidium parvum and Giardia lamblia. Appl Environ Microbiol. 2004;70(1):597–598. | |
Liandris E, Gazouli M, Andreadou M, Sechi LA, Rosu V, Ikonomopoulos J. Detection of pathogenic mycobacteria based on functionalized quantum dots coupled with immunomagnetic separation. PLoS ONE. 2011;6(5):220026. | |
Wang L, Wu C-S, Fan X, Mustapha A. Detection of Escherichia coli O157:H7 and Salmonella in ground beef by a bead-free quantum dot- facilitated isolation method. Int J Food Microbiol. 2012;156(1):83–87. | |
Brandt SM, Paulin SM. Quantifying colonization potential of enterohaemorrhagic Escherichia coli O157:H7 using bovine in vitro organ culture and immunofluorescent staining. Foodborne Pathog Dis. 2012;9(12):1064–1070. | |
Zhao Y, Ye M, Chao Q, Jia N, Ge Y, Shen H. Simultaneous detection of multifood-borne pathogenic bacteria based on functionalized quantum dots coupled with immunomagnetic separation in food samples. J Agric Food Chem. 2009;57:517–524. | |
Zhu H, Sikora U, Ozcan A. Quantum dot detection of Escherichia coli using a cell-phone. Analyst. 2012;137:2541–2544. | |
Goldman ER, Clapp AR, Anderson GP, et al. Multiplexed toxin analysis using four colors of quantum dot fluororeagents. Anal Chem. 2004;76(3):684–688. | |
Yang Y, Xu F, Xu H, et al. Magnetic nano-beads based separation combined with propidium monoazide treatment and multiplex PCR assay for simultaneous detection of viable Salmonella typhimurium, Escherichia coli O157:H7 and Listeria monocytogenes in food products. Food Microbiol. 2013;34(2):418–424. | |
Fu J, Park B, Siragusa G, et al. An Au/Si hetero-nanorod-based biosensor for Salmonella detection. Nanotechnology. 2008;19:155502. | |
Narsaiah K, Jha SN, Bhardwaj R, Sharma R, Kumar R. Optical biosensors for food quality and safety assurance: a review. J Food Sci Technol. 2012;49(4):383–406. | |
Davis D, Gup X, Musavi L, Lin C-S, Chen S-Z, Wu VCH. Gold nanoparticle-modified carbon electrode biosensor for the detection of Listeria monocytogenes. Ind Biotechnol. 2013;9(1):31–36. | |
Dudak FC, Boyaci IH. Development of an immunosensor based on surface plasmon resonance for enumeration of Escherichia coli in water samples. Food Res Int. 2007;40(7):803–807. | |
Song Y, Li W, Duan Y, Li Z, Deng L. Nicking enzyme-assisted biosensor for Salmonella enteritidis detection based on fluorescence resonance energy transfer. Biosens Bioelectron. 2014;55(0):400–404. | |
McIntyre L, Hudson JA, Billington C, Withers H. Biocontrol of foodborne bacteria. In: McElhattan A, Sobral P, editors. Novel Technologies in Food Science – Their Impact on Products, Consumer Trends and Environment. New York, USA: Springer Publishers; 2012:183–204. | |
Ackermann H-W. Bacteriophage classification. In: Kutter EM, Sulakvelidze A, editors. Bacteriophages; Biology and Applications. Boca Raton, FL, USA: CRC Press; 2005:67–89. | |
Rohwer F. Global phage diversity. Cell. 2003;113(2):141. | |
Chang TC, Ding HC, Chen S. A conductance method for the identification of Escherichia coli O157:H7 using bacteriophage AR1. J Food Prot. 2002;65(1):12–17. | |
Amorim LRP, Silva JGL, Gibbs PA, Teixeira PC. Application of an impedimetric technique for the detection of lytic infection of Salmonella spp. by specific phages. Int J Microbiol. 2009;2009:259456. | |
Jassim SAA, Griffiths MW. Evaluation of a rapid microbial detection method via phage lytic amplification assay coupled with Live/Dead fluorochromic stains. Lett Appl Microbiol. 2007;44(6):673–678. | |
Stewart GS, Jassim SA, Denyer SP, Newby P, Linley K, Dhir VK. The specific and sensitive detection of bacterial pathogens within 4 h using bacteriophage amplification. J Appl Microbiol. 1998;84(5):777–783. | |
Stewart LD, Foddai A, Elliott CT, Grant IR. Development of a novel phage-mediated immunoassay for the rapid detection of viable Mycobacterium avium subsp. paratuberculosis. J Appl Microbiol. 2013;115(3):808–817. | |
Rees JC, Voorhees KJ. Simultaneous detection of two bacterial pathogens using bacteriophage amplification coupled with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun Mass Spectrom. 2005;19(19):2757–2761. | |
Pierce CL, Rees JC, Fernández FM, Barr JR. Viable Staphylococcus aureus quantitation using 15N metabolically labeled bacteriophage amplification coupled with a multiple reaction monitoring proteomic workflow. Mol Cell Proteomics. 2012;11(1):M111.012849. | |
Ripp S, Jegier P, Johnson CM, Brigati JR, Satyler GS. Bacteriophage-amplified bioluminescent sensing of Escherichia coli O157:H7. Anal Bioanal Chem. 2008;391:507–514. | |
Loessner MJ, Rudolf M, Scherer S. Evaluation of luciferase reporter bacteriophage A511::luxAB for detection of Listeria monocytogenes in contaminated foods. Appl Environ Microbiol. 1997;63(8):2961–2965. | |
Waddell TE, Poppe C. Construction of mini-Tn10luxABcam/ Ptac-ATS and its use for developing a bacteriophage that transduces bioluminescence to Escherichia coli O157:H7. FEMS Microbiol Lett. 2000;182(2):285–289. | |
Chen J, Griffiths MW. Salmonella detection in eggs using Lux+ bacteriophages. J Food Prot. 1996;59(9):908–914. | |
Pagotto F, Brovko L, Griffiths MW. Phage-mediated detection of Staphylococcus aureus and Escherichia coli 0157:H7 using bioluminescence. Paper presented at: the International Dairy Federation Symposium on the Bacteriological Quality of Raw Milk; 1996; Wolfpassing, Austria. | |
Irwin P, Gehring A, Tu SI, Brewster J, Fanelli J, Ehrenfeld E. Minimum detectable level of Salmonellae using a binomial-based bacterial ice nucleation detection assay (BIND). J AOAC Int. 2000;83(5):1087–1095. | |
Hagens S, de Wouters T, Vollenweider P, Loessner MJ. Reporter bacteriophage A511::celB transduces a hyperthermostable glycosidase from Pyrococcus furiosus for rapid and simple detection of viable Listeria cells. Bacteriophage. 2011;1(3):143–151. | |
Willford J, Goodridge LD. An integrated assay for rapid detection of Escherichia coli O157:H7 on beef samples. Food Prot Trends. 2008;28:468–474. | |
Tanji Y, Furukawa C, Na S-H, Hijikata T, Miyanaga K, Unno H. Escherichia coli detection by GFP-labeled lysozyme-inactivated T4 bacteriophage. J Biotechnol. 2004;114:11–20. | |
Hoang HA, Abe M, Nakasaki K. A novel colorimetric method for the detection of Escherichia coli using cytochrome c peroxidase-encoding bacteriophage. FEMS Microbiol Lett. 2014;352(1):97–103. | |
Goodridge L, Chen J, Griffiths M. The use of a fluorescent bacteriophage assay for detection of Escherichia coli O157:H7 in inoculated ground beef and raw milk. Int J Food Microbiol. 1999;47(1–2):43–50. | |
Edgar R, McKinstry M, Hwang J, et al. High-sensitivity bacterial detection using biotin-tagged phage and quantum-dot nanocomplexes. Proc Natl Acad Sci. 2006;103(13):4841–4845. | |
Chai Y, Wikle HC, Wang Z, et al. Design of a surface-scanning coil detector for direct bacteria detection on food surfaces using a magnetoelastic biosensor. J Appl Phys. 2013;114(104504). | |
Park M-K, Li S, Chin BA. Detection of Salmonella Typhimurium grown directly on tomato surface using phage-based magnetoelastic biosensors. Food and Bioprocess Technology. 2013;6(3):682–689. | |
Chai Y, Li S, Horikawa S, Park M-K, Vodyanoy V, Chin BA. Rapid and sensitive detection of Salmonella typhimurium on eggshells by using wireless biosensors. J Food Prot. 2012;75(4):631–636. | |
Lakshmanan RS, Guntupalli R, Hu J, Petrenko VA, Barbaree JM, Chin BA. Detection of Salmonella typhimurium in fat free milk using a phage immobilized magnetoelastic sensor. Sens Actuators B Chem. 2007;126(2):544–550. | |
Arya SK, Singh A, Naidoo R, Wu P, McDermott MT, Evoy S. Chemically immobilized T4-bacteriophage for specific Escherichia coli detection using surface plasmon resonance. Analyst. 2011;136(3):486–492. | |
Tawil N, Sacher E, Mandeville R, Meunier M. Surface plasmon resonance detection of E. coli and methicillin-resistant S. aureus using bacteriophages. Biosens Bioelectron. 2012;37(1):24–29. | |
Balasubramanian S, Sorokulova IB, Vodyanoy VJ, Simonian AL. Lytic phage as a specific and selective probe for detection of Staphylococcus aureus: a surface plasmon resonance spectroscopic study. Biosens Bioelectron. 2007;22(6):948–955. | |
Laube T, Cortés P, Llagostera M, Alegret S, Pividori M. Phagomagnetic immunoassay for the rapid detection of Salmonella. Appl Microbiol Biotechnol. 2013;98(4):1–11. | |
Fernandes E, Martins VC, Nóbrega C, et al. A bacteriophage detection tool for viability assessment of Salmonella cells. Biosens Bioelectron. 2014;52(0):239–246. | |
Singh A, Arutyunov D, McDermott MT, Szymanski CM, Evoy S. Specific detection of Campylobacter jejuni using the bacteriophage NCTC 12673 receptor binding protein as a probe. Analyst. 2011;136(22):4780–4786. | |
Walcher G, Stessl B, Wagner M, Eichenseher F, Loessner MJ, Hein I. Evaluation of paramagnetic beads coated with recombinant Listeria phage endolysin-derived cell-wall-binding domain proteins for separation of Listeria monocytogenes from raw milk in combination with culture-based and real-time polymerase chain reaction-based quantification. Foodborne Pathog Dis. 2010;7(9):1019–1024. | |
Tolba M, Ahmed MU, Tlili C, Eichenseher F, Loessner MJ, Zourob M. Bacteriophage endolysin-based electrochemical impedance biosensor for the rapid detection of Listeria cells. Analyst. 2012;137(24):5749–5756. | |
Gill CO. Visible contamination on animals and carcasses and the microbiological contamination of meat. J Food Prot. 2004;67(2):413–419. | |
Oosterom J, Notermans CS, Karman H, Engels GB. Origin and prevalence of Campylobacter jejuni in poultry processing. J Food Prot. 1983;46(4):339–344. | |
Feldhusen F. The role of seafood in bacterial foodborne diseases. Microbes Infect. 2000;2(13):1651–1660. | |
Jordan D, McEwen SA, Lammerding AM, McNab WB, Wilson JB. Pre-slaughter control of Escherichia coli O157 in beef cattle: a simulation study. Prev Vet Med. 1999;41(1):55–74. | |
Bardina C, Spricigo DA, Cortés P, Llagostera M. Significance of the bacteriophage treatment schedule in reducing Salmonella colonization of poultry. Appl Environ Microbiol. 2012;78(18):6600–6607. | |
Wong CL, Sieo CC, Tan WS, et al. Evaluation of a lytic bacteriophage, Φ st1, for biocontrol of Salmonella enterica serovar Typhimurium in chickens. Int J Food Microbiol. 2014;172(0):92–101. | |
Waseh S, Hanifi-Moghaddam P, Coleman R, et al. Orally administered P22 phage tailspike protein reduces Salmonella colonization in chickens: prospects of a novel therapy against bacterial infections. PLoS ONE. 2010;5(11):e13904. | |
JaneŽ N, Loc-Carrillo C. Use of phages to control Campylobacter spp. J Microbiol Methods. 2013;95(1):68–75. | |
Kittler S, Fischer S, Abdulmawjood A, Glünder G, Klein G. Effect of bacteriophage application on Campylobacter jejuni loads in commercial broiler flocks. Appl Environ Microbiol. 2013;79(23):7525–7533. | |
Carvalho CM, Gannon BW, Halfhide DE, et al. The in vivo efficacy of two administration routes of a phage cocktail to reduce numbers of Campylobacter coli and Campylobacter jejuni in chickens. BMC Microbiol. 2010;20:232. | |
Chekabab SM, Paquin-Veillette J, Dozois CM, Harel J. The ecological habitat and transmission of Escherichia coli O157:H7. FEMS Microbiol Lett. 2013;341:1–12. | |
Raya RR, Oot RA, Moore-Maley B, et al. Naturally resident and exogenously applied T4-like and T5-like bacteriophages can reduce Escherichia coli O157:H7 levels in sheep guts. Bacteriophage. 2011;1:15–24. | |
Niu YD, Xu Y, McAllister TA, et al. Comparison of fecal versus rectoanal mucosal swab sampling for detecting Escherichia coli O157:H7 in experimentally inoculated cattle used in assessing bacteriophage as a mitigation strategy. J Food Prot. 2008;71(4):691–698. | |
Coffey B, Rivas L, Duffy G, Coffey A, Ross RP, McAuliffe O. Assessment of Escherichia coli O157:H7-specific bacteriophage e11/2 and e4/1c in model broth and hide environments. Int J Food Microbiol. 2011;147(3):188–194. | |
Luftig R, Pelon W. Bacteriophage biodepuration of Vibrio vulnificus containing shellfish. First International Conference on Shellfish Restoration. Hilton Head Island, SC, USA; 1996. | |
Pelon W, Luftig RB, Johnston KH. Vibrio vulnificus load reduction in oysters after combined exposure to Vibrio vulnificus-specific bacteriophage and to an oyster extract component. J Food Prot. 2005;68(6):1188–1191. | |
Oliveira M, Viñas I, Colàs P, Anguera M, Usall J, Abadias M. Effectiveness of a bacteriophage in reducing Listeria monocytogenes on fresh-cut fruits and fruit juices. Food Microbiol. 2014;38:137–142. | |
Rossi LPR, Almeida RCC, Lopes AL, Figueiredo ACL, Ramos MPP, Almeida PF. The occurrence of Listeria spp. in Brazilian fresh sausage and control of Listeria monocytogenes using bacteriophage P100. Food Control. 2011;22(6):954–958. | |
Soni KA, Nannapaneni R, Hagens S. Reduction of Listeria monocytogenes on the surface of fresh channel catfish fillets by bacteriophage Listex P100. Foodborne Pathog Dis. 2010;7(4):427–434. | |
Soni KA, Nannapaneni R. Bacteriophage significantly reduces Listeria monocytogenes on raw salmon fillet tissue. J Food Prot. 2010;73(1):32–38. | |
Soni KA, Desai M, Oladunjoye A, Skrobot F, Nannapaneni R. Reduction of Listeria monocytogenes in queso fresco cheese by a combination of listericidal and listeriostatic GRAS antimicrobials. Int J Food Microbiol. 2012;155(1–2):82–88. | |
Carlton RM, Noordman WH, Biswas B, de Meester ED, Loessner MJ. Bacteriophage P100 for control of Listeria monocytogenes in foods: genome sequence, bioinformatic analysis, oral toxicity study, and application. Regul Toxicol Pharmacol. 2005;43:301–312. | |
Guenther S, Huwyler D, Richard S, Loessner MJ. Virulent bacteriophage for efficient biocontrol of Listeria monocytogenes in ready-to-eat foods. Appl Environ Microbiol. 2009;75(1):93–100. | |
Holck A, Berg J. Inhibition of Listeria monocytogenes in cooked ham by virulent bacteriophages and protective cultures. Appl Environ Microbiol. 2009;75(21):6944–6946. | |
Bigot B, Lee WJ, McIntyre L, et al. Control of Listeria monocytogenes growth in a ready-to-eat poultry product using a bacteriophage. Food Microbiol. 2011;28:1448–1452. | |
Hudson JA, Billington C, Cornelius AJ, et al. Use of a bacteriophage to inactivate Escherichia coli O157:H7 on beef. Food Microbiol. 2013;36(1):14–21. | |
Hudson JA, Billington C, Wilson T, On SLW. Effect of phage and host concentration on the inactivation of Escherichia coli O157:H7 on cooked and raw beef. Food Sci Technol Int. Epub November 27, 2013. | |
Carter CD, Parks A, Abuladze T, et al. Bacteriophage cocktail significantly reduces Escherichia coli O157: H7 contamination of lettuce and beef, but does not protect against recontamination. Bacteriophage. 2012;2(3):178–185. | |
Viazis S, Akhtar M, Feirtag J, Diez-Gonzalez F. Reduction of Escherichia coli O157:H7 viability on leafy green vegetables by treatment with a bacteriophage mixture and trans-cinnamaldehyde. Food Microbiol. 2011;28(1):149–157. | |
Sharma M, Patel JR, Conway WS, Ferguson S, Sulakvelidze A. Effectiveness of bacteriophages in reducing Escherichia coli O157:H7 on fresh-cut cantaloupes and lettuce. J Food Prot. 2009;72(7):1481–1485. | |
Abuladze T, Li M, Menetrez MY, Dean T, Senecal A, Sulakvelidze A. Bacteriophages reduce experimental contamination of hard surfaces, tomato, spinach, broccoli and ground beef by Escherichia coli O157:H7. Appl Environ Microbiol. 2008;74(20):6230–6238. | |
Kang H-W, Kim J-W, Jung T-S, Woo G-J. wksl3, a new biocontrol agent for Salmonella enterica serovars Enteritidis and Typhimurium in foods: characterization, application, sequence analysis, and oral acute toxicity study. Appl Environ Microbiol. 2013;79(6):1956–1968. | |
Hooton SPT, Atterbury RJ, Connerton IF. Application of a bacteriophage cocktail to reduce Salmonella typhimurium U288 contamination on pig skin. Int J Food Microbiol. 2011;15:157–163. | |
Ye J, Kostrzynska M, Dunfield K, Warriner K. Control of Salmonella on sprouting mung bean sprouts and alfalfa seeds by using a biocontrol preparation based on antagonistic bacteria and lytic phages. J Food Prot. 2010;73(1):9–17. | |
Higgins JP, Higgins SE, Guenther KL, et al. Use of a specific bacteriophage treatment to reduce Salmonella in poultry. Poult Sci. 2005;84:1141–1145. | |
Bigwood T, Hudson JA, Billington C, Carey-Smith GV, Heinemann JA. Phage inactivation of foodborne pathogens on cooked and raw meat. Food Microbiol. 2008;25:400–406. | |
Goode D, Allen VM, Barrow PA. Reduction of experimental Salmonella and Campylobacter contamination of chicken skin by application of lytic bacteriophage. Appl Environ Microbiol. 2003;69(8):5032–5036. | |
Modi R, Hirvi Y, Hill A, Griffiths MW. Effect of phage on survival of Salmonella enteritidis during manufacture and storage of cheddar cheese made from raw and pasteurised milk. J Food Prot. 2001;64:927–933. | |
Atterbury RJ, Connerton PL, Dodd CER, Rees CED, Connerton IF. Application of host-specific bacteriophages to the surface of chicken skin leads to a reduction in recovery of Campylobacter jejuni. Appl Environ Microbiol. 2003;69(10):6302–6306. | |
Lee W-J, Billington C, Hudson JA, Heinemann JA. Isolation and characterization of phages infecting Bacillus cereus. Lett Appl Microbiol. 2011;52:456–464. | |
Bueno E, García P, Martínez B, Rodríguez A. Phage inactivation of Staphylococcus aureus in fresh and hard-type cheeses. Int J Food Microbiol. 2012;158(1):23–27. | |
Hudson JA, Bigwood T, Premaratne A, Billington C, Horn B. Potential to use UV-inactivated bacteriophages to control foodborne pathogens. Foodborne Pathog Dis. 2010;7(6):687–693. | |
Anany H, Chen W, Pelton R, Griffiths MW. Biocontrol of Listeria monocytogenes and Escherichia coli O157:H7 in meat by using phages immobilized on modified cellulose membranes. Appl Environ Microbiol. 2011;77(18):6379–6387. | |
Cademartiri R, Anany H, Gross I, Bhayani R, Griffiths M, Brook MA. Immobilization of bacteriophages on modified silica particles. Biomaterials. 2010;31:1904–1910. | |
Korehi R, Kadla J. Incorporating pre-encapsulated bacteriophage in electrospun fibres. Paper presented at: 16th International Symposium on Wood, Fiber and Pulping Chemistry; 2011; Tianjin, People’s Republic of China. | |
Solanki K, Grover N, Downs P, et al. Enzyme-based listericidal nanocomposites. Sci Rep. 2013;3:1584. | |
Rashidi L, Khosravi-Darani K. The applications of nanotechnology in food industry. Crit Rev Food Sci Nutr. 2011;51:723–730. | |
Nedovic V, Kalusevic A, Manojlovic V, Levic S, Bugarski B. An overview of encapsulation technologies for food applications. Procedia Food Sci. 2011;1:1806–1815. | |
Malheiros PDS, Daroit DJ, da Silveira NP, Brandelli A. Effect of nanovesicle-encapsulated nisin on growth of Listeria monocytogenes in milk. Food Microbiol. 2010;27:175–178. | |
Imran M, Revol-Junelles A, René N, et al. Microstructure and physico-chemical evaluation of nano-emulsion-based antimicrobial peptides embedded in bioactive packaging films. Food Hydrocoll. 2012;29:407–419. | |
Food Safety Authority of Ireland. The Relevance for Food Safety of Applications of Nanotechnology in the Food and Feed Industries: Food Additives, Contaminants and Chemical Residues. Dublin, Ireland: Food Safety Authority of Ireland; 2008. | |
Appendini P, Hothchkiss JH. Review of antimicrobial food packaging. Innovative Food Science and Engineering Technologies. 2002;3:113–126. | |
de Azeredo HMC. Antimicrobial nanostructures in food packaging. Trends Food Sci Technol. 2013;30:56–69. | |
Dutta PK, Tripathi S, Mehrotra GK, Dutta J. Perspectives for chitosan based antimicrobial films in food applications. Food Chem. 2009;1114:1173–1182. | |
Aider M. Chitosan application for active bio-based films production and potential in the food industry: review. Food Sci Technol. 2010;43:837–842. | |
Hosseini SF, Zandi M, Rezaei M, Farahmandghavi F. Two-step method for encapsulation of oregano essential oil in chitosan nanoparticles: preparation, characterization and in vitro release study. Carbohydr Polym. 2013;95:50–56. | |
Xing K, Chen WG, Li YY, et al. Antibacterial activity of oleoyl-chitosan nanoparticles: a novel antibacterial dispersion system. Carbohydr Polym. 2008;74:114–120. | |
Alishahi A. Antibacterial effect of chitosan nanoparticle loaded with nisin for prolonged effect. J Food Saf. 2014;34:111–118. | |
Liu H, Du Y, Wang X, Sun L. Chitosan kills bacteria through cell membrane damage. Int J Food Microbiol. 2004;95:147–155. | |
Xing K, Chen XG, Liu CS, Cha DS, Park HJ. Oleoyl-chitosan nanoparticles inhibits Escherichia coli and Staphylococcus aureus by damaging the cell memebrane and putative binding to extracellular or intracellular targets. Int J Food Microbiol. 2009;132:127–133. | |
Abdollahi M, Rezaei M, Farzi G. A novel active bionanocomposite film incorporating rosemary essential oil and nanoclay into chitosan. J Food Eng. 2012;(111):343–350. | |
Rhim J, Park H, Ha C. Bio-nanocomposites for food packaging applications. Prog Polym Sci. 2013;38:1629–1652. | |
Li L, Lin S-L, Deng L, Zu Z-G. Potential use of chitosan nanoparticles for oral delivery of DNA vaccine in black seabreem Acanthopagrus schlegelii Bleeker to protect from Vibrio parahaemolyticus. J Fish Dis. 2013;36:987–995. |
 © 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.