Back to Journals » Cancer Management and Research » Volume 16
PERP May Affect the Prognosis of Lung Adenocarcinoma by Inhibiting Apoptosis
Authors Liu Z, Han S, Luo Y, Zhao Z, Ni L, Chai L, Tang H
Received 14 November 2023
Accepted for publication 29 February 2024
Published 20 March 2024 Volume 2024:16 Pages 199—214
DOI https://doi.org/10.2147/CMAR.S443490
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Antonella D'Anneo
Zhongxiang Liu,1,* Shuhua Han,2,* Yuhong Luo,3,* Zhangyan Zhao,4 Lingyu Ni,5 Linlin Chai,6 Haicheng Tang4
1Department of Pulmonary and Critical Care Medicine, the Yancheng Clinical College of Xuzhou Medical University, The First People’s Hospital of Yancheng, the First Affiliated Hospital of Jiangsu Vocational College of Medicine, Yancheng, 224000, People’s Republic of China; 2Department of Respiratory and Critical Care Medicine, Zhongda Hospital, School of Medicine, Southeast University, Nanjing, Jiangsu, 210009, People’s Republic of China; 3College of Life Science and Technology, Guangxi University, Nanning, 530004, People’s Republic of China; 4Department of Respiratory and Critical Care Medicine, Shanghai Public Health Clinical Center, Fudan University, Shanghai, 201508, People’s Republic of China; 5China School of Life Sciences, Nanjing University, Nanjing, Jiangsu, 210046, People’s Republic of China; 6Department of Pathology, The Yancheng Clinical College of Xuzhou Medical University, The First People’s Hospital of Yancheng, The First Affiliated Hospital of Jiangsu Vocational College of Medicine, Yancheng, 224000, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Linlin Chai, Department of Pathology, The Yancheng Clinical College of Xuzhou Medical University, The First People’s Hospital of Yancheng, Yancheng, Jiangsu, People’s Republic of China, Email [email protected] Haicheng Tang, Department of Respiratory and Critical Care Medicine, Shanghai Public Health Clinical Center, Fudan University, 2901, Caolang Road, Jinshan District, Shanghai, 201508, People’s Republic of China, Email [email protected]
Background: PERP, a member of the peripheral myelin protein gene family, is a new therapeutic target in cancer. The relationships between PERP and immune cell infiltration in lung cancer have not been studied. Therefore, the role of PERP in the tumour microenvironment (TME) of lung cancer needs to be further explored.
Methods: In this study, we explored the association between PERP expression and clinical characteristics by analysing data from the TCGA database. Cox regression and Kaplan‒Meier methods were used to investigate the relationship between the expression of PERP and overall survival in patients with lung adenocarcinoma (LUAD). The relationship between PERP expression and the degree of infiltration of specific immune cell subsets in LUAD was evaluated using the TIMER database and GEPIA. We also performed GO enrichment analysis and KEGG enrichment analysis to reveal genes coexpressed with PERP using the Coexpedia database. Finally, we verified the expression and function of PERP in LUAD tissues and the A549 cell line by RT‒PCR, Western blot, CCK-8, IHC, and wound healing assays. The mouse model was used to study the in vivo effects of PERP.
Results: According to our results, PERP expression was significantly higher in LUAD tissues and associated with the clinical characteristics of the disease. Survival was independently associated with PERP in LUAD patients. We further verified that PERP might regulate B-cell infiltration in LUAD to affect the prognosis of LUAD. To identify PERP-related signalling pathways in LUAD, we performed a genome-aggregation analysis (GSEA) between low and high PERP expression datasets. LUAD cells express higher levels of PERP than paracarcinoma cells, and PERP inhibits the proliferation and metastasis of A549 cells through apoptosis.
Conclusion: PERP may affect the prognosis of lung adenocarcinoma by inhibiting apoptosis and is associated with immune cell infiltration.
Keywords: PERP, lung adenocarcinoma, apoptosis, prognosis
Introduction
Approximately 85% of lung cancer deaths are caused by non-small cell lung cancer (NSCLC).1,2 There are several types of NSCLC, including adenocarcinomas, squamous cell carcinomas, and others.3 Adenocarcinomas are among the most common types of lung cancer, so they are very important in terms of treatment and outcome. Especially for lung adenocarcinomas, immunotherapy has provided hope for treating lung cancer.4,5 However, the tumour microenvironment (TME), which is based on enhancing immunological defences against cancer, may impair the efficacy of immunotherapy.6–8 An increasing number of researchers are studying how the TME affects tumour prognosis.
The TME consists of a variety of cellular and acellular components, including immune cells, endothelial cells, fibroblasts, and extracellular matrix.9–11 Alteration of immune cell infiltration in the TME may induce immune checkpoint expression and lead to novel immunotherapeutic strategies to prolong the life of patients with cancers.12–14 As an example, infiltration of T cells increases the expression of programmed death ligand 1 (PD-L1) and improves PD-1 inhibitor efficacy.15,16 These results demonstrate that immune cell infiltration in the TME is critical in lung cancer treatment and prognosis.
The PERP gene belongs to the peripheral myelin protein 22-kDa (PMP22) gene family; multiple members of the PMP22 gene family may be targets for cancer treatment.17 The PERP effector regulates apoptosis and epithelial-mesenchymal transition through signalling pathways dependent upon p53 or other signalling pathways.18 PERP mRNA N6 adenosine methylation mediated by METTL14 leads to pancreatic cancer progression, which offers a new hope for treatment.19 Another study showed that PERP variants can have different antioxidant activities by modulating p53 stability and influencing cancer risk.20 Additionally, some researchers believe that PERP gives rise to tumour suppression by inhibiting the growth and metastasis of tumours. For example, breast cancer growth can be inhibited by PERP by inhibiting TGF-β signalling.21 In addition, no studies have examined the relationship between PERP and immune cell infiltration in lung cancer. Therefore, the role of PERP in the TME of lung cancer needs to be further explored.
In the present study, we explored the relationship between PERP and immune cell infiltration in the TME of lung adenocarcinoma and its impact on the prognosis of lung adenocarcinoma through multiple bioinformatics analyses, which were demonstrated experimentally.
Materials and Methods
Analysis of Gene Expression and Clinical Data
In March 2021, TCGA database gene expression data (mRNA, HTSeq-FPKM) and clinical data were downloaded for 535 lung adenocarcinoma tissue samples and 59 normal tissue samples (https://portal.gdc.cancer.gov/repository).
Analysis of PERP Expression and Survival in Bioinformatics
Through analysis of the TCGA database, the expression levels of PERP were compared between lung adenocarcinoma and normal samples. We analysed these data to determine whether PERP expression is associated with lung adenocarcinoma clinical features. Patients with lung adenocarcinoma were analysed using Cox regression analysis and Kaplan‒Meier methods to determine whether PERP expression is related to overall survival.
Analysis of Enriched Gene Sets
GSEA can determine whether two biological states show significant and consistent differences in terms of a set of genes defined a priori.22 To investigate the significant survival differences between high and low PERP lung cancer samples, we performed GSEA to obtain an ordered list of all genes based on their association with PERP expression. Sequences of 1000 genomes were used in each analysis. Phenotypic markers were assessed using PERP expression levels. Each phenotype was classified according to normal P values and normalized enrichment scores (NES).
Analysis of TIMER Databases
Based on the TIMER database (https://cistrome.shinyapps.io/timer/), we evaluated the relationship between PERP expression and specific subsets of immune cells infiltrating lung adenocarcinoma.23 Using Kaplan‒Meier curve analysis, we investigated whether gene expression or immunosuppression influenced patient survival.24
Analysis of PERP-Associated Immunomodulators
Through TISIDB (http://cis.hku.hk/TISIDB/), heterogeneous types of data are integrated to understand how tumours interact with the immune system, through which we can extract PERP-related immunomodulators.25 We further analysed the immunostimulants and immunosuppressants significantly associated with PERP expression by the Spearman correlation test.
Lung Cancer Explorer Online Analysis
Through Lung Cancer Explorer (http://lce.biohpc.swmed.edu/lungcancer/), data from TCGA, GEO, and other databases were combined to perform meta-analyses of PERP gene expression and survival in lung adenocarcinomas.26
Database Analysis for GEPIA
The GEPIA database (http://gepia.cancer-pku.cn/) offers a way to assess the relationship between PERP expression and markers associated with tumour infiltration by immune cells.27
Coexpedia Analysis
The Coexpedia database (http://www.coexpedia.org/) was used to analyse the coexpressed genes of PERP. The database is derived from the GEO dataset based on human and mouse samples.28 A filtering network was established for PERP as medical subject words. Gene scores are based on the sum of the edge weights (logarithmic likelihoods) for all connected genes.
The Metascape online database integrating more than 40 bioinformatics studies could create protein‒protein interaction networks by extracting enriched annotations and identifying enriched pathways.29 Using Metascape’s online database, we examined the GO and KEGG enrichment for PERP-coexpressed genes.
Cell Lines and Culture Conditions
With RPMI-1640 medium containing 10% FBS, penicillin/streptomycin and 5% carbon dioxide, the human A549 cell line obtained from Shanghai Institute of Cell Biology was cultured at 37°C.
Reverse Transcription-Polymerase Chain Reaction (RT‒PCR) is a Technique for Isolating and Quantifying RNA
Total RNA was isolated from cells or tissues by RNAiso Plus reagent (TaKaRa Biotechnology, 9109) according to the manufacturer’s protocol, and cDNA synthesis was processed by a reverse transcription kit according to the manufacturer’s instructions (ABclonal, RK20429). Two-dimensional PCR was conducted on an Applied Biosystems 7500 Cycler (Applied Biosystems) using four-element SYBR Taq (ABclonal, RK21203), 0.4 μL, 10 μM primer mixtures, and 2 μL of cDNA. With the 2−ΔΔCt analysis method, the relative expression level was determined, in which 18S was used as an internal standard. All reactions were repeated three times. The primer sequences for the target genes were as follows: PERP forward 5’-CTTCACCCTTCATGCCAACC-3’, reverse 5′-GCCAATCAGGATAATCGTGGCT-3’; 18S forward 5’-AACCCGTTGAACCCCATT-3’, reverse 5’-CCATCCAATCGGTAGTAGCG-3’.
Western Blot Analysis
The cell lysate was prepared with RIPA (Beyotime, P0013B) buffer with complete protease inhibitors. After that, a Pierce BCA protein assay kit (23225 from Thermo Scientific) was used to measure the protein concentrations. Approximately 20 mg of total protein from each group was separated by sodium dodecyl sulfate‒polyacrylamide gel electrophoresis and transferred to a polyvinylidene fluoride membrane. Incubations were performed overnight at 4 °C with a 1:1000 dilution of the primary antibody. A rabbit anti-human antibody against PERP (ABclonal, A5937) was used as the secondary antibody. Antibodies against caspase 3 (CST, #9662) were used as primary antibodies. Antibodies against cleaved caspase 3 and cleaved PARP (CST, #5625) were used as primary antibodies. A secondary antibody (mouse or rabbit human, Santa Cruz Biotech, 1:4000) was incubated for an hour at room temperature on the membranes.
Immunohistochemistry (IHC)
Using immunohistochemistry, we identified 20 lung adenocarcinomas and adjacent tissues from Yancheng First People’s Hospital. Slides with formalin-fixed paraffin embedded (FFPE) tissues were deparaffinized and deproteinized. Sodium citrate buffer was placed in the samples for 30 minutes (microwave heating) for antigenic repair. For the elimination of endogenous peroxidase activity, samples were preincubated with 3% H2O2. After 30 minutes of blocking with goat serum, anti-PERP (ABclonal, A5937) was incubated overnight at 4 °C with the sections. A goat anti-rabbit IgG biotinylated with streptavidin-biotin conjugated with horseradish peroxidase (HRP) was incubated at 37 °C for one hour. After that, the signals were visualized using a diaminobenzidine kit (ZSGB-BIO), and slides were counterstained with haematoxylin. Counterstaining was performed using haematoxylin. An experienced pathologist blinded to the clinical information scored, evaluated, and analysed IHC staining.
Construction of Plasmids and Transfection
Transfection of A549 cells with PERP short hairpin RNA (shRNA) and control vectors used as retroviral silencing vectors was performed. The shRNA targeting sequence of PERP was as follows: CGTGACCAAATATCCTGAAAT, CGTGAAGTACACCCAGACCTT.
Proliferation Analysis of Cells
The effect of knocking down PERP on A549 cell proliferation was examined using the CCK-8 assay. After plasmid transfection, bladder cancer cells were seeded at 1000 per well in 96-well plates. For at least five replicate wells, CCK-8 was added for 1–4 hours at 37 °C. With a 450 nm plate reader, the OD was calculated. An EdU staining plate was used to count the cells after resuspension in RPMI-1640 with 10% FBS containing EdU (10 μmol/L). Two hours were spent culturing cells (5×105/well). Afterwards, we stained samples with C10310-1 (RiboBio) according to standard procedures. In addition to staining the sections with DAPI, we sealed the sections with a quencher to prevent fluorescence from escaping. The EdU-positive cell count was calculated by multiplying the EdU-positive cell count by the EdU-positive cell count + EdU-negative cell count by 100%.
Analyses of Cell Apoptosis
Three kinds of shPERPs were transfected into cells: shCONT, shPERP-1 and shPERP-2. Annexin V-APC/PI (KGA1030-100) binding buffer was added to the cell suspension after the cells had been treated for 72 hours. Finally, the cells were analysed using flow cytometry (Beckman Coulter Cytoflex; Beckman, USA).
Wound-Healing Assay
Wound-healing assays were used to measure the migration of A549 cells. Our procedure consisted of inflicting a uniform wound into each plate after the growing cell layers reached confluence and then washing the wound layer with PBS to remove any cell debris. During the experiment, images were taken at the beginning and after 24 hours, and the migration rate of the cells was determined.
In vivo Experiments in Nude Mice
Female Nude mouse were purchased from the Experimental Animal Center of Nanjing Medical University and raised under pathogen free conditions. In order to establish a lung cancer xenograft model, A549 cells stably interfered with PERP were suspended in 100 mL PBS and subcutaneously inoculated into the ribs and abdomen of nude mice. The nude mice were randomly divided into two groups (3 in each group). Measure and record the length (L) and width (W) of the tumor, and calculate the tumor volume (V): V= (L × W2) × 0.5.
Statistical Analysis
Wilcoxon tests were used to examine relationships between PERP expression and clinicopathological features, and visualizations were made using R (version 3.6.1). TCGA database data were analysed with univariate Kaplan‒Meier (KM) and Cox regression analyses to determine overall survival (OS). The effects of PERP expression along with other clinicopathological variables were compared using multivariate Cox regression models. Expression levels at higher and lower levels were determined based on median values of PERP. For statistical analysis, SPSS version 20.0 (SPSS, Chicago, IL, USA) and GraphPad Prism 7.0 (GraphPad, San Diego, CA, USA) were used. Student’s t-tests and one-way ANOVAs were used. P< 0.05 was considered statistically significant.
Results
Expression of PERP and Clinical Characteristics of Lung Adenocarcinoma
Based on information from the TISIDB database, we compared tumours and normal tissues in various cancers for PERP mRNA expression (Figure 1). Adenocarcinoma of the lung and its adjacent tissues showed differential expression of PERP. According to the TCGA database, the expression of PERP mRNA in lung adenocarcinoma was examined. As shown in Figure 2A, the mRNA expression of PERP was significantly upregulated in lung adenocarcinoma tissues compared with normal tissues (P= 4.152e−16). The results were verified by meta-analysis in the LCE database for reliability and authenticity (hazard ratio (HR)=1.13, Figure 2B).
 |
Figure 1 Online analysis of the TISIDB confirmed that PERP is expressed in various cancers as well as normal tissues. *P<0.05, ***P<0.001. |
According to the results of the univariate logistic regression analysis (Table 1), PERP expression differed significantly between the T stage (T2 vs T1, P=0.04) and N stage (N1 vs N0, P=0.01; N2 vs N0, P=0.04). In lymph node involvement with multifactorial aggravation, PERP expression was increased.
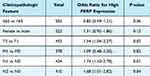 |
Table 1 Association Between PERP Expression and Clinical Characteristics |
Accordingly, PERP expression was significantly elevated in lung adenocarcinoma tissues and associated with the clinical characteristics of lung adenocarcinomas, possibly due to the P53 signalling pathway.
Survival Analysis of PERP in Lung Adenocarcinoma
For further assessment of PERP and its prognostic value, in TCGA cases, high and low expression levels of PERP were determined by median values, and survival analysis was performed. According to the Kaplan‒Meier analysis, the overall survival time of the low-PERP group was better than that of the high-PERP group (P=5.279e-05; Figure 3A). Time-dependent ROC analysis showed that the AUC at 1 year, 3 years, and 5 years was 0.661 0.627 and 0.595, suggesting that patients with lung adenocarcinomas might benefit from PERP expression (Figure 3B). Meta-analysis of LCE database data confirmed that PERP is an effective prognosticator (hazard ratio (HR)= 1.34, Figure 3C). As shown in Table 2, Cox regression analyses were used to identify prognostic factors. Based on a univariate analysis, pathological stage, tumour stage (T), lymph node stage (N), and PERP expression influenced OS for patients with lung adenocarcinoma. As shown in Table 2 and Figure 3D, PERP was an independent prognostic factor according to multivariate Cox regression analysis.
 |
Table 2 Cox Regression Analysis of the Prognostic Factors in Lung Adenocarcinoma Univariate Analysis |
Lung Adenocarcinoma Immune Cell Infiltration and PERP Expression
The expression level of PERP and tumour purity were not significantly correlated (R= −0.025, P=5.79e-01), as shown in Figure 4A. However, in lung adenocarcinoma, the expression was significantly associated with B-cell infiltration (R= −0.189, P=2.79e-05), macrophage infiltration (R = 0.113, P = 1.27e-02), and neutrophil infiltration (R = 0.117, P = 9.94e-03) but not CD4+ T-cell infiltration (R= −0.035, P= 4.42e-01), CD8+ T-cell infiltration (R = 0.075, P=9.90e-02) or dendritic cell infiltration (R = 0.071, P =1.17e-01). In addition, using the Timer database, we generated Kaplan‒Meier plots to examine the correlation between immune cell infiltration and prognosis for lung adenocarcinomas. Adenocarcinoma prognosis was negatively influenced by B-cell infiltration (P=0) and dendritic cell infiltration (P=0.048) (Figure 4B), suggesting that PERP is implicated in regulating the infiltration of B cells by lung adenocarcinomas. In the TIMER database, we explored the correlation between the number of somatic copies and the amount of immune infiltration. Deleting PERP at the chromosome arm level significantly decreased the infiltration levels of CD8+ T cells, CD4+ T cells, macrophages, neutrophils and dendritic cells (P<0.05, Figure 4C).
In addition, based on our analysis of PERP expression and immune markers in immune cells, such as B cells, CD8+ T cells, CD4+ T cells, neutrophils, monocytes, and dendritic cells, we observed a correlation (Table 3). The expression of PERP was found to have a negative correlation with B-cell immunomarkers in lung adenocarcinoma but not in paracancerous tissues. PERP expression was negatively correlated with CD8+ T cells and monocyte immunomarkers in the adjacent tissues of lung adenocarcinoma but not in the cancerous tissue itself. Negative correlations were also found between this molecule and most dendritic cell immunomarkers in both cancerous and paracancerous tissues. Additionally, PERP was negatively related to PDCD1 in lung adenocarcinoma. Additionally, no correlation was found between this molecule and antibodies against CD4+ T cells.
 |
Table 3 Correlation Between PERP Expression and Immune Markers of Immune Cells in GEPIA |
In summary, PERP regulated B-cell infiltration in lung adenocarcinoma, which might affect the prognosis of lung adenocarcinoma.
PERP-Related Signalling Pathways in Lung Adenocarcinoma
We conducted genome-aggregation analysis (GSEA) on low and high PERP expression datasets to identify PERP-related signalling pathways in lung adenocarcinoma. The GSEA results showed significant differences in the enrichment of the MSigDB collection (h.all.v7.2.symbols.gmt) (nominal P < 0.05 OR FDR q < 0.25). The normalized enrichment score (NES) was used to determine the most significantly enriched signalling pathway, which was plotted in a multi-GSEA enrichment map (Figure 5A). The multi-GSEA enrichment map showed that the cell cycle, DNA repair, other enzymes involved in drug metabolism, glucosphingolipid biosynthesis of Lactobacillus and NeoLactobacillus series, nucleotide excision repair, P53 signalling pathway, pentose and glucuronate interconversions and pyrimidine metabolism were associated with the high expression of PERP. In addition, vascular smooth muscle contraction was associated with low expression of PERP. Therefore, these PERP-related signalling pathways might be involved in the development of lung adenocarcinoma.
Coexpression Analysis of PERP
Coexpression networks for PERP were predicted using the Coexpedia database, resulting in 437 associated genes, which were subsequently analysed for GO enrichment (Figure 5B and C). As shown in Figure 5C, the genes targeting membrane cotranslation proteins, epidermal development, cell‒cell junctions, desmosomes, cell adhesion molecule binding, and keratinocyte proliferation were enriched. We also performed KEGG enrichment analysis, which showed that the genes of ribosomes, oestrogen signalling pathways, arrhythmogenic right ventricular cardiomyopathy (ARVC), adherent junction, Hippo signalling pathway, TGF-β signalling pathway, sphingolipid signalling pathway, pathogenic E. coli infection, Vibrio cholerae infection, Alzheimer’s disease, cancer pathway, P53 signalling pathway, and cytochrome P450 metabolism of heterobiomaterial were enriched (Figure 5D).
Identification of the Expression and Function of PERP in LUAD
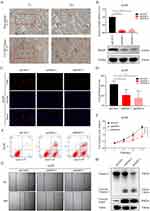
Our bioinformatics analysis was further verified by collecting 20 pairs of LUAD tissue and adjacent tissue samples. As part of the investigation of the expression of PERP protein in LUAD tissues and adjacent tissues, immunohistochemistry (IHC) was performed. Comparing adjacent tissues with LUAD tissues, Figure 6A shows significantly increased PERP staining signals. The fact that PERP levels are associated with a poor prognosis in LUAD patients prompted us to investigate whether this protein is involved in the functioning of oncogenes. After silencing PERP, the relative mRNA and protein levels of PERP were decreased in A549 cells (Figure 6B). As shown in Figure 6C–E, according to the EdU and Cell Counting Kit 8 (CCK-8) assays, A549 cells with PERP knockdown had significantly decreased viability and proliferation. As determined by flow cytometry, transfection of A549 cells with sh-PERP increased apoptosis (Figure 6F). We also assessed the migratory properties of LUAD cells by wound-healing assays. In this study, simulating PERP significantly impaired the migration of A549 cells (Figure 6G). Additionally, knocking down PERP induced increased levels of apoptosis in LUAD cells, marked by elevated levels of cleaved caspase-3 and cleaved PARP (Figure 6H).
Interference with PERP to Inhibit Tumor Growth in vivo
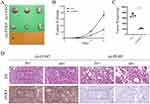
To further evaluate the in vivo effect of PERP interference on NSCLC cells, A549 cells with stable PERP interference were inoculated into BALB/C thymic free mice. As shown in the Figure 7A, tumors formed by cells interfered with PERP were smaller than those formed by control cells, and the tumor volume and weight were lower (Figure 7B and C). The expression of PERP in transplanted tumor tissue was analyzed using IHC (Figure 7D).
Discussion
Immunotherapy has shown promising results in lung cancer patients, and new diagnostic and treatment strategies have been suggested based on the analysis of the characteristics of the TME and molecular signature of immune-related genes.14,30,31 New treatments have decreased lung adenocarcinoma incidence and mortality, but it remains one of the most prevalent cancers.32 Therefore, it is necessary to search for new diagnostic methods and immunotherapeutic targets. In this study, we found that the expression of PERP was significantly increased in lung adenocarcinoma tissues compared to normal tissues and closely associated with the clinical characteristics of lung adenocarcinoma. We also demonstrated that high expression of PERP was an independent risk factor for lung adenocarcinoma through Cox regression analysis. In addition, it was found that the infiltration of immune cells was closely related to the expression of PERP, especially the deletion of PERP at the chromosome arm level. Furthermore, we identified PERP-related signalling pathways that might play an important role in the development of lung adenocarcinoma.
Recently, PERP was found to be a member of the four transmembrane proteins (PMP22 family) involved in the regulation of apoptosis and intercellular adhesion.18 The role of PERP in cancer has been controversial since it is a p53 effector. High expression of PERP has been shown to be associated with lymph node metastasis of papillary thyroid carcinoma.33 However, other studies have suggested that PERP inhibits tumour growth and progression by promoting apoptosis.34,35 In our study, meta-analysis of the LCE database showed increased PERP expression in lung adenocarcinoma tissues compared to paracancerous tissues. In a multivariate logistic regression, the level of PERP expression was correlated with the T and N phases of development. Furthermore, the survival analysis showed that patients with lung adenocarcinoma who expressed low levels of PERP were more likely to survive. Therefore, in lung adenocarcinomas, we believe that PERP can promote tumour growth.
Immune cells have been found to infiltrate NSCLC in a variety of ways, with CD4+ T cells, CD8+ T cells, and B cells being the most commonly found.36,37 Researchers have found a close association between immune cells and survival in lung cancer patients. In most studies, there is an association between T-cell infiltration and lung cancer prognosis. For example, there is a link between T-reg cells and poor prognosis in tumours and CD4+ T-cell infiltration with good prognosis in non-small cell lung cancer.38,39 It has been reported that patients with NSCLC who have high densities of B cells have an improved survival rate. Depending on how they function, B cells are involved in antitumour immunity either by capturing tumour antigens and presenting them to T cells or by producing antibodies against tumour antigens.40–42 As major regulators of immune and cancerous responses, dendritic cells are associated with better outcomes in NSCLC.43 This study found that PERP regulates B-cell infiltration in lung adenocarcinoma, which may affect patient prognosis. Increased regulation of PERP can activate caspase to induce the apoptosis of B cells, so we speculate that PERP may affect adenocarcinoma of the lung with B-cell infiltration by inducing apoptosis, which requires further verification.44 Our study revealed a significant decrease in the infiltration levels of various immune cells, including B cells, upon deletion of PERP at the chromosomal arm level via the SCNA module in the TIMER database. Patients with lung adenocarcinoma who have B-cell infiltration have a higher chance of surviving. This result suggests a worse prognosis in lung adenocarcinoma with the deletion of PERP at the chromosomal arm level. In other cancer studies, it was found that PERP variants can also affect patient prognosis.19,20 We therefore concluded that PERP variants may increase the risk of cancer by affecting immune cell infiltration.
The immunomodulatory function of PERP was also confirmed by the analysis of PERP-related immunoenhancers and immunosuppressants. One of the most promising approaches today for predicting biomarkers and antitumour therapies is immune checkpoint-based selection.45–47 Tumour progression is affected by immune cell infiltration in the TME, and lung adenocarcinomas are associated with high immune cell infiltration.48 Therefore, lung adenocarcinomas may be prognosticated based on PERP-influenced B-cell infiltration as well as treatable through PERP therapy.
Furthermore, PERP expression is associated with various signalling pathways, including the cell cycle, DNA repair, and P53. Coexpression analysis of PERP shows enrichment for genes involved in the Hippo, TGF-β, and P53 signalling pathways, among others. Targeting P53 signalling and modifying relevant protein expression are important for NSCLC proliferation and invasion.49 P53 signalling pathways are involved in host immunity surveillance and tumour immunity evasion, leading to targeted inhibitors of these pathways in active immunotherapy.50 Lung adenocarcinomas inhibit the Hippo signalling pathway, and relieving this inhibition might improve the disease’s prognosis.51 In lung adenocarcinomas, TGF-β signalling can activate fibroblasts and promote immune escape.52,53 Therefore, we need to further explore which signalling pathway is involved in B-cell infiltration regulated by PERP in lung adenocarcinoma.
In summary, we explored whether adenocarcinoma prognosis is influenced by PERP expression. There is an independent risk factor for lung adenocarcinoma associated with high expression of PERP in lung adenocarcinoma tissues. Apoptosis was found to affect A549 cell proliferation and metastasis as well. In addition, there is a correlation between PERP expression and immune cell infiltration, particularly B cells, and adenocarcinomas of the lung are regulated by the tumour microenvironment.
Data Sharing Statement
The original source data and material of first author will be available upon reasonable request.
Ethics Approval and Consent to Participate
The studies involving human participants were reviewed and approved by the Ethics Committee of the First People’s Hospital of Yancheng (No. 2023-K-061). Animal experiments have been approved by the Ethics Committee of Jiangsu Medical Vocational College (No. XMLL-2022-854). The research content of experimental animals complies with the “Regulations on the Management of Experimental Animals” issued by the National Science and Technology Commission. The patients provided written informed consent to participate in this study. Our study complies with the Declaration of Helsinki.
Acknowledgments
We are grateful for the data provided based on the professional network platform and thank all authors who participated in this study for their cooperation.
Funding
This work was supported by Shanghai Municipal Health Commission (No. 202040332); Jiangsu Provincial Health Commission, Elderly Health Research Project (No. LKM2022075); Shanghai Public Health Clinical Center (No. KY-GW-2021-16).
Disclosure
The authors declare that they have no competing interests to declare in this work.
References
1. Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi:10.3322/caac.21660
2. Cao W, Chen HD, Yu YW, Li N, Chen Q. Changing profiles of cancer burden worldwide and in China: a secondary analysis of the global cancer statistics 2020. Chin Med J. 2021;134(7):783–791. doi:10.1097/CM9.0000000000001474
3. Shi Y, Chen W, Li C, et al. Clinicopathological characteristics and prediction of cancer-specific survival in large cell lung cancer: a population-based study. J Thorac Dis. 2020;12(5):2261–2269. doi:10.21037/jtd.2020.04.24
4. Saito M, Suzuki H, Kono K, Takenoshita S, Kohno T. Treatment of lung adenocarcinoma by molecular-targeted therapy and immunotherapy. Surg Today. 2018;48(1):1–8. doi:10.1007/s00595-017-1497-7
5. Ruiz-Cordero R, Devine WP. Targeted therapy and checkpoint immunotherapy in lung cancer. Surg Pathol Clin. 2020;13(1):17–33. doi:10.1016/j.path.2019.11.002
6. Frankel T, Lanfranca MP, Zou W. The role of tumor microenvironment in cancer immunotherapy. Adv Exp Med Biol. 2017;1036:51–64. doi:10.1007/978-3-319-67577-0_4
7. Wei F, Wang D, Wei J, et al. Metabolic crosstalk in the tumor microenvironment regulates antitumor immunosuppression and immunotherapy resisitance. Cell Mol Life Sci. 2021;78(1):173–193. doi:10.1007/s00018-020-03581-0
8. Cheng N, Bai X, Shu Y, Ahmad O, Shen P. Targeting tumor-associated macrophages as an antitumor strategy. Biochem Pharmacol. 2021;183:114354. doi:10.1016/j.bcp.2020.114354
9. Fridman WH. The tumor microenvironment: prognostic and theranostic impact. Recent advances and trends. Semin Immunol. 2020;48:101416. doi:10.1016/j.smim.2020.101416
10. Wang JJ, Lei KF, Han F. Tumor microenvironment: recent advances in various cancer treatments. Eur Rev Med Pharmacol Sci. 2018;22(12):3855–3864. doi:10.26355/eurrev_201806_15270
11. Arneth B. Tumor microenvironment. Medicina. 2019;56(1):15. doi:10.3390/medicina56010015
12. Gajewski TF, Schreiber H, Fu YX. Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol. 2013;14(10):1014–1022. doi:10.1038/ni.2703
13. Zhang J, Endres S, Kobold S. Enhancing tumor T cell infiltration to enable cancer immunotherapy. Immunotherapy. 2019;11(3):201–213. doi:10.2217/imt-2018-0111
14. Fridman WH, Zitvogel L, Sautes-Fridman C, Kroemer G. The immune contexture in cancer prognosis and treatment. Nat Rev Clin Oncol. 2017;14(12):717–734. doi:10.1038/nrclinonc.2017.101
15. Ayers M, Lunceford J, Nebozhyn M, et al. IFN-gamma-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest. 2017;127(8):2930–2940. doi:10.1172/JCI91190
16. Bodor JN, Boumber Y, Borghaei H. Biomarkers for immune checkpoint inhibition in non-small cell lung cancer (NSCLC). Cancer. 2020;126(2):260–270. doi:10.1002/cncr.32468
17. Wang YW, Cheng HL, Ding YR, Chou LH, Chow NH. EMP1, EMP 2, and EMP3 as novel therapeutic targets in human cancer. Biochim Biophys Acta Rev Cancer. 2017;1868(1):199–211. doi:10.1016/j.bbcan.2017.04.004
18. Roberts O, Paraoan L. PERP-ing into diverse mechanisms of cancer pathogenesis: regulation and role of the p53/p63 effector PERP. Biochim Biophys Acta Rev Cancer. 2020;1874(1):188393. doi:10.1016/j.bbcan.2020.188393
19. Wang M, Liu J, Zhao Y, et al. Upregulation of METTL14 mediates the elevation of PERP mRNA N(6) adenosine methylation promoting the growth and metastasis of pancreatic cancer. Mol Cancer. 2020;19(1):130. doi:10.1186/s12943-020-01249-8
20. Liao CY, Yang SF, Wu TJ, et al. Novel function of PERP-428 variants impacts lung cancer risk through the differential regulation of PTEN/MDM2/p53-mediated antioxidant activity. Free Radic Biol Med. 2021;167:307–320. doi:10.1016/j.freeradbiomed.2021.02.017
21. Xia L, Xiao X, Liu WL, et al. Coactosin-like protein CLP/Cotl1 suppresses breast cancer growth through activation of IL-24/PERP and inhibition of non-canonical TGFbeta signaling. Oncogene. 2018;37(3):323–331. doi:10.1038/onc.2017.342
22. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi:10.1073/pnas.0506580102
23. Li T, Fan J, Wang B, et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77(21):e108–e110. doi:10.1158/0008-5472.CAN-17-0307
24. Dai Y, Qiang W, Lin K, Gui Y, Lan X, Wang D. An immune-related gene signature for predicting survival and immunotherapy efficacy in hepatocellular carcinoma. Cancer Immunol Immunother. 2021;70(4):967–979. doi:10.1007/s00262-020-02743-0
25. Ru B, Wong CN, Tong Y, et al. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics. 2019;35(20):4200–4202. doi:10.1093/bioinformatics/btz210
26. Cai L, Lin S, Girard L, et al. LCE: an open web portal to explore gene expression and clinical associations in lung cancer. Oncogene. 2019;38(14):2551–2564. doi:10.1038/s41388-018-0588-2
27. Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47(W1):W556–W560. doi:10.1093/nar/gkz430
28. Yang S, Kim CY, Hwang S, et al. COEXPEDIA: exploring biomedical hypotheses via co-expressions associated with medical subject headings (MeSH). Nucleic Acids Res. 2017;45(D1):D389–D396. doi:10.1093/nar/gkw868
29. Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523. doi:10.1038/s41467-019-09234-6
30. Ling B, Huang Z, Huang S, Qian L, Li G, Tang Q. Microenvironment analysis of prognosis and molecular signature of immune-related genes in lung adenocarcinoma. Oncol Res. 2021;28(6):561–578. doi:10.3727/096504020X15907428281601
31. Qin N, Li Y, Wang C, et al. Comprehensive functional annotation of susceptibility variants identifies genetic heterogeneity between lung adenocarcinoma and squamous cell carcinoma. Front Med. 2021;15(2):275–291. doi:10.1007/s11684-020-0779-4
32. Myers DJ, Wallen JM. Lung Adenocarcinoma. Treasure Island (FL): StatPearls; 2021.
33. Zhai T, Muhanhali D, Jia X, Wu Z, Cai Z, Ling Y. Identification of gene co-expression modules and hub genes associated with lymph node metastasis of papillary thyroid cancer. Endocrine. 2019;66(3):573–584. doi:10.1007/s12020-019-02021-9
34. Holmes BJ, von Eyben R, Attardi LD, Kong CS, Le QT, Nathan CO. Pilot study of loss of the p53/p63 target gene PERP at the surgical margin as a potential predictor of local relapse in head and neck squamous cell carcinoma. Head Neck. 2020;42(11):3188–3196. doi:10.1002/hed.26358
35. Chen B, Li Z, Feng Y, Wu X, Xu Y. Myocardin-related transcription factor A (MRTF-A) mediates doxorubicin-induced PERP transcription in colon cancer cells. Biochem Biophys Res Commun. 2018;503(3):1732–1739. doi:10.1016/j.bbrc.2018.07.106
36. Stankovic B, Bjorhovde HAK, Skarshaug R, et al. Immune Cell Composition in Human Non-small Cell Lung Cancer. Front Immunol. 2018;9:3101. doi:10.3389/fimmu.2018.03101
37. Fridman WH, Remark R, Goc J, et al. The immune microenvironment: a major player in human cancers. Int Arch Allergy Immunol. 2014;164(1):13–26. doi:10.1159/000362332
38. Menetrier-Caux C, Gobert M, Caux C. Differences in tumor regulatory T-cell localization and activation status impact patient outcome. Cancer Res. 2009;69(20):7895–7898. doi:10.1158/0008-5472.CAN-09-1642
39. Hiraoka K, Miyamoto M, Cho Y, et al. Concurrent infiltration by CD8+ T cells and CD4+ T cells is a favourable prognostic factor in non-small-cell lung carcinoma. Br J Cancer. 2006;94(2):275–280. doi:10.1038/sj.bjc.6602934
40. Remark R, Becker C, Gomez JE, et al. The non-small cell lung cancer immune contexture. A major determinant of tumor characteristics and patient outcome. Am J Respir Crit Care Med. 2015;191(4):377–390. doi:10.1164/rccm.201409-1671PP
41. Chen J, Tan Y, Sun F, et al. Single-cell transcriptome and antigen-immunoglobin analysis reveals the diversity of B cells in non-small cell lung cancer. Genome Biol. 2020;21(1):152. doi:10.1186/s13059-020-02064-6
42. Yuen GJ, Demissie E, Pillai S. B lymphocytes and cancer: a love-hate relationship. Trends Cancer. 2016;2(12):747–757. doi:10.1016/j.trecan.2016.10.010
43. Dieu-Nosjean MC, Antoine M, Danel C, et al. Long-term survival for patients with non-small-cell lung cancer with intratumoral lymphoid structures. J Clin Oncol. 2008;26(27):4410–4417. doi:10.1200/JCO.2007.15.0284
44. Iijima K, Yamada H, Miharu M, et al. ZNF 385 B is characteristically expressed in germinal center B cells and involved in B-cell apoptosis. Eur J Immunol. 2012;42(12):3405–3415. doi:10.1002/eji.201242530
45. Darvin P, Toor SM, Sasidharan Nair V, Elkord E. Immune checkpoint inhibitors: recent progress and potential biomarkers. Exp Mol Med. 2018;50(12):1–11. doi:10.1038/s12276-018-0191-1
46. Li B, Chan HL, Chen P. Immune checkpoint inhibitors: basics and challenges. Curr Med Chem. 2019;26(17):3009–3025. doi:10.2174/0929867324666170804143706
47. Domagala-Kulawik J. New frontiers for molecular pathology. Front Med. 2019;6:284. doi:10.3389/fmed.2019.00284
48. Zuo S, Wei M, Wang S, Dong J, Wei J. Pan-cancer analysis of immune cell infiltration identifies a Prognostic Immune-Cell Characteristic Score (ICCS) in lung adenocarcinoma. Front Immunol. 2020;11:1218. doi:10.3389/fimmu.2020.01218
49. Jiang W, Hou L, Wei J, et al. Hsa-miR-217 inhibits the proliferation, migration, and invasion in non-small cell lung cancer cells via targeting SIRT1 and P53/KAI1 signaling. Balkan Med J. 2020;37(4):208–214. doi:10.4274/balkanmedj.galenos.2020.2019.9.91
50. Cui Y, Guo G. Immunomodulatory function of the tumor suppressor p53 in host immune response and the tumor microenvironment. Int J Mol Sci. 2016;17. doi:10.3390/ijms17111942
51. An Y, Zhang Q, Li X, Wang Z, Li Y, Tang X. Upregulated microRNA miR-21 promotes the progression of lung adenocarcinoma through inhibition of KIBRA and the Hippo signaling pathway. Biomed Pharmacother. 2018;108:1845–1855. doi:10.1016/j.biopha.2018.09.125
52. Kang JI, Kim DH, Sung KW, et al. p62-induced cancer-associated fibroblast activation via the Nrf2-ATF6 pathway promotes lung tumorigenesis. Cancers. 2021;13(4):864. doi:10.3390/cancers13040864
53. Jang HR, Shin SB, Kim CH, et al. PLK1/vimentin signaling facilitates immune escape by recruiting Smad2/3 to PD-L1 promoter in metastatic lung adenocarcinoma. Cell Death Differ. 2021;28(9):2745–2764. doi:10.1038/s41418-021-00781-4
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.