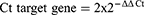Back to Journals » The Application of Clinical Genetics » Volume 16
Copy Number Variation in the GSTM1 and GSTT1 Genes and the Risk of Liver Cirrhosis in Eastern Ethiopia
Authors Mekuria AN , Seyoum T , Alemayehu DH, Abebe M, Nedi T , Abula T, Gong YY, Engidawork E
Received 17 August 2023
Accepted for publication 17 October 2023
Published 20 October 2023 Volume 2023:16 Pages 171—179
DOI https://doi.org/10.2147/TACG.S435852
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Martin Maurer
Abraham Nigussie Mekuria,1 Tamrayehu Seyoum,2 Dawit Hailu Alemayehu,2 Markos Abebe,2 Teshome Nedi,1 Tefera Abula,1 Yun Yun Gong,3 Ephrem Engidawork1
1Department of Pharmacology and Clinical Pharmacy, School of Pharmacy, College of Health Sciences, Addis Ababa University, Addis Ababa, Ethiopia; 2Department of Biotechnology and Bioinformatics, Armauer Hansen Research Institute, Addis Ababa, Ethiopia; 3School of Food Science and Nutrition, University of Leeds, Leeds, UK
Correspondence: Ephrem Engidawork, Email [email protected]
Background: Polymorphisms in glutathione S-transferase M1 (GSTM1) and T1 (GSTT1) can cause an entire gene deletion. The current methodology can accurately identify GSTM1 and GSTT1 copy number variants (CNVs), which may shed light on the true contribution of each gene copy to the cellular detoxification process and disease risk. Because liver cirrhosis is becoming a critical worldwide health issue, this study determined the CNVs of GSTM1 and GSTT1 and their relationship to the risk of liver cirrhosis.
Methods: In this study, we compared 106 patients with liver cirrhosis to 104 healthy controls. Real-time PCR was used to identify the CNVs of GSTM1 and GSTT1. Logistic and linear regression models were used to estimate the relationship between liver cirrhosis and clinical chemistry variables with the CNVs, respectively.
Results: In 3.3% of the study participants, > 2 copies of the GSTM1 or GSTT1 genes were detected. GSTT1 carriers had a significantly lower risk of liver cirrhosis (p< 0.05) compared with individuals who had homozygous deletion (adjusted odds ratio (AOR) = 0.47; 95% CI: 0.25, 0.86). This risk reduction was significant (p< 0.05) in patients with a single copy of the GSTT1 gene (AOR = 0.48; 95% CI: 0.25, 0.91). Those with ≥ 2 copies of combined GSTM1 and GSTT1 also had a significantly (p< 0.05) lower risk of developing liver cirrhosis compared with double null genotypes (AOR = 0.38; 95% CI: 0.16, 0.91, p trend < 0.001). Moreover, ≥ 2 copies of combined GSTM1 and GSTT1 genes were associated with a substantial decrease in alanine amino transferase (ALT) and aspartate aminotransferase (AST) levels, respectively.
Conclusion: A single copy number of GSTT1, and ≥ 2 copies of combined GSTM1 and GSTT1 genes were associated with a reduced risk of liver cirrhosis in Ethiopians. These findings underscore the importance of gene–environment interactions in the multifactorial development of liver cirrhosis.
Keywords: liver cirrhosis, copy number variation, glutathione S-transferase genes, Ethiopia
Introduction
Glutathione S-transferase (GST) enzymes are generally known to protect cells from oxidative stress due to metabolic products of endogenous and exogenous chemicals.1 GST’s biochemical defense mechanisms include both the reduction of organic hydroperoxides, which contribute to oxidative stress, and the conjugation of electrophilic chemicals with glutathione, which facilitates their transport out of the cell.2,3 There are 16 genes coding for the cytosolic form of GST enzymes in humans and are classified into 7 classes: alpha (GSTA), mu (GSTM), pi (GSTP), theta (GSTT), omega (GSTO), and zeta (GSTZ).1 These genes are known to be highly polymorphic,4 which can result in altered detoxification and oxidative stress defense capacity in certain tissues, particularly the liver, which has a central metabolic role.5,6
Oxidative stress is thought to be the primary cause of liver injury and progression to liver cirrhosis as well as hepatocellular carcinoma (HCC) associated with aflatoxin B1 (AFB1) exposure.7,8 In our recent study, we found a strong association between AFB1 exposure and liver cirrhosis.9 AFB1 is mainly metabolized by CYP3A4 into the reactive free radical AFB1-exo-8, 9-epoxide.10 This intermediate metabolite is typically detoxified by conjugation with glutathione through the action of GST enzymes, mainly GSTM1 and GSTT1 encoded by the GSTM1 and GSTT1 gene, respectively.11 Copy number variation (CNV) has been ascribed to these genes and individuals with zero copy number (null genotype) of GSTM1 and/or GSTT1 are more prone to develop HCC associated with AFB1 exposure.12,13 However, previous reports failed to distinguish between heterozygous and homozygous bearers of the non-deleted alleles. Hence, the present study aimed at investigating GSTT1 and GSTM1 CNVs and the risk for liver cirrhosis in Eastern Ethiopia.
Materials and Methods
Study Subjects
This is a follow-up of a case–control study conducted from January 2020 – July 2021 to assess the risk factors associated with liver cirrhosis in the Internal Medicine Unit of Hiwot Fana Comprehensive Specialized University Hospital (HFCSUH), Harar, Eastern Ethiopia.9 In brief, the study consisted of 127 confirmed cases of liver cirrhosis and 253 controls without any history or clinical evidence of liver diseases. Written consent from the participants was obtained after explaining aims of the study and a detailed questionnaire was filled out for each case and control by a trained interviewer. A subset of 210 individuals, including all cases (106), for whom blood samples were available, and an equivalent number of controls (104) were randomly selected and included in this genetic association study.
Blood Collection
Two mL of blood was collected in an EDTA tube for genomic DNA extraction. Additional 3–5 mL blood was also taken in a serum separator tube for clinical chemistry tests. Whole blood samples collected for genomic DNA extraction were stored at −80°C until transport and analysis at the Armauer Hansen Research Institute, Addis Ababa, Ethiopia.
DNA Extraction
Genomic DNA was isolated from whole blood using PureLink™ Genomic DNA Mini Kit (Invitrogen) according to the manufacturer’s instructions. DNA quality and quantity was assessed using NanoDrop 2000 spectrophotometer (ThermoFisher Scientific, USA). Moreover, 10% of the samples were randomly selected and agarose gel electrophoresis with the aid of a gel imaging system, Gel Doc (Bio-Rad), was performed to ensure quality of the extracted DNA. The genomic DNA samples were stored at −20°C until analysis.
GSTM1 and GSTT1 Copy Number Analysis
Copy number determination was carried out by a Bio-Rad CFX96 Touch Real-Time System. The reaction mixture (10 uL) was composed of FAM-labeled 20x TaqManTM Copy Number Assay kit (AB Applied Biosystems) for GSTM1 (Hs02595872_cn) or GSTT1 (Hs01731033_cn), VIC-labeled 20X TaqManTM Reference Assay kit (AB Applied Biosystems: 4403326), TaqPathTM ProAmpTM Master Mix (Applied Biosystems), nuclease-free water and 10 ng genomic DNA template. Thermocycling was initiated at 95°C for 10 min followed by 40 cycles of 15 sec of denaturation at 95°C and 60-sec annealing-extension at 60°C.
To ensure quality, each target assay was done in the same PCR as RNaseP. The amplification of RNaseP and GST genes in the same sample adjusted DNA concentration differences between samples and ensured that no false-negative GST*0/0 genotypes were created due to PCR or pipetting failure or inadequate DNA concentrations in the original sample. The genotyping was done blind to the subject’s case or control status. The samples were run twice. In addition, each run contained a no-template control to rule out any contamination. The number of amplification cycles necessary for the fluorescent signal to reach the threshold and surpass the background level is known as the cycle threshold (Ct) value in the qPCR experiment. All samples were quantified in duplicates and average Ct values were normalized to RNase P (the reference gene with copy number 2). Copy number estimation was conducted by the ∆∆Ct method as follows:
where ΔΔCt= ΔCtGST sample – ΔCtRNase P and ΔCt was the average Ct value of 2 repeated measurements of the sample. The result obtained from the 2−ΔΔCt calculation was multiplied by 2 because the target genes are known to be diploid.14,15
Clinical Chemistry
Liver function test (LFT) including aspartate transaminase (AST), alanine transaminase (ALT), alkaline phosphatase (ALP), serum bilirubin (total), albumin, and serum creatinine was determined using Cobas C-311 fully automated closed clinical chemistry analyzer (Roche/Hitachi Cobas C-311, Roche diagnostics GmbH, Mannheim, Germany).
Statistical Analysis
Categorical variables were expressed as frequencies and percentages, while continuous variables were expressed as median (IQR). The difference between cases and controls in terms of categorical and continuous variables was tested using χ2 or Fisher’s exact test and the nonparametric Mann–Whitney-U test, respectively. Using unconditional logistic regression, odds ratios (ORs) were estimated for GSTM1 and GSTT1 genotypes, comparing groups defined by the gene copy number coded as a categorical variable (0, 1, or ≥2 copies), null, and carriers (1, 2 or more copies). Given that gender and age are biological risk factors for liver cirrhosis, we adjusted the OR for these variables as potential confounders. The association between clinical chemistry tests and GSTM1 and GSTT1 copy numbers and genotypes was also investigated using linear regression. Since the Shapiro–Wilk test revealed a non-normal distribution of the clinical chemistry data, we used the log-transferred data during regression analysis. The adjustment was made using the potential confounders (age and gender). P<0.05 was considered statistically significant. All statistical analyses were performed using Statistical Package for Social Sciences (SPSS) version 26.0.
Ethical Considerations
This study was conducted following the Declaration of Helsinki. Ethical approval was obtained from both the Institutional Review Board of the College of Health Sciences, Addis Ababa University (Protocol No. 064/19/SOP and Reference No. CHS/RTTD/229/2020) and the National Ethical Review Committee of the Ministry of Science and Higher Education (Reference No. 04/246/680/21). Written consent was obtained following provision of information regarding the study’s objectives, benefits, and risks to participants. Confidentiality and anonymity were ensured by restricting data access and removing identifiers, respectively.
Results
Sociodemographic and Clinical Characteristics
Table 1 shows characteristics of the study participants. Males accounted for a higher number of cases (67%) than controls (52%) and we found statistically significant differences (p<0.05) regarding gender but not age between the two groups. There was a significant (p<0.05) difference between cases and controls in terms of all clinical chemistry parameters (Table 1).
 |
Table 1 Sociodemographic and Clinical Chemistry Characteristics of the Study Participants, Eastern Ethiopia, 2020/21 (N=210) |
Copy Number Variation
Figure 1 depicts CNV of the study participants. The GSTM1 null genotype was more common in cases (0.45; 95% CI: 0.36, 0.55) than controls (0.33; 95% CI: 0.24, 0.43), with an overall frequency of 0.39 (95% CI: 0.32, 0.46) in the study population. Similarly, the frequency of GSTT1 null genotype was significantly higher (p<0.05) in cases (0.40; 95% CI: 0.30, 0.50) than controls (0.25; 95% CI: 0.17, 0.34), with an overall frequency of 0.32 (95% CI: 0.26, 0.39). The overall percentage of double null GSTM1 and GSTT1 genotypes was 13% (95% CI: 0.09, 0.19) and relatively more common in cases (16%; 95% CI: 0.10, 0.24) than controls (11%; 95% CI: 0.05, 0.18). Moreover, 3.3% (95% CI: 0.01, 0.07) of the study participants had >2 copies of the GSTM1 or GSTT1 genes, each accounting for 1.9% (95% CI: 0.01, 0.05) and 1.4% (95% CI: 0.01, 0.04) for GSTT1), respectively.
 |
Figure 1 Frequency of GSTM1 and GSTT1 copy numbers among the study participants, Eastern Ethiopia, 2020/21 (n=210). |
Association Studies
As shown in Table 2, GSTT1 carriers had a significantly (p<0.05) lower risk of liver cirrhosis than individuals with null genotypes (AOR=0.47; 95% CI: 0.25, 0.86). Further analysis also revealed that patients with one copy of the GSTT1 gene had a significantly (p<0.05) lower risk of liver cirrhosis than patients with null genotypes (AOR=0.48; 95% CI: 0.25, 0.91). Moreover, the risk of liver cirrhosis was markedly (p<0.05) decreased in those patients who had ≥2 combined GSTM1 and GSTT1 copies compared with patients who had a dual null genotype (AOR=0.38; 95% CI: 0.16, 0.91). There was also a significant gene dose relationship (p-trend < 0.001).
 |
Table 2 Risk of Liver Cirrhosis in Relation to GSTM1 and GSTT1 Genes Copy Numbers and Genotypes, Eastern Ethiopia, 2020/21 (N=210) |
We further investigated whether GSTM1 and GSTT1 copy numbers and genotypes indirectly contribute to the levels of clinical chemistry parameters as shown in Table 3. Consequently, the ALT level was reduced by 31% (β= 0.69; 95% CI: 0.48, 0.99; p<0.05) in cases with ≥2 copies of GSTM1 in comparison with null genotypes (Table 3). Moreover, AST level was decreased by 39% (β= 0.61; 95% CI: 0.41, 0.90; p<0.05) in cases having ≥2 copies of GSTT1 than those with a null genotype. Similarly, AST level showed a significant (p<0.05) reduction in cases who were carriers of the GSTT1 genotype compared to null genotypes (β= 0.76; 95% CI: 0.58, 0.98) (Table 3).
 |
Table 3 Linear Regression of the Association Between ALT, AST, GSTM1, and GSTT1 Copy Numbers and Genotypes, Eastern Ethiopia, 2020/21 (N=210) |
Discussion
Previous studies suggest that polymorphisms in genes encoding Phase II metabolizing enzymes, such as GSTM1 and GSTT1 may be linked to an increased vulnerability to various disorders associated with oxidative stress.16,17 To the best of our knowledge, this is the first study showing the contribution of the exact CNV of GSTM1 and GSTT1 to liver cirrhosis. In our study, both GSTM1 and GSTT1 null genotypes were more frequently detected in liver cirrhosis patients than controls and the overall frequency was 0.39 and 0.32, respectively. Furthermore, 13% of the study population had double GSTM1 and GSTT1 null genotypes, whereas 3.3% (1.9% for GSTM1 and 1.4% for GSTT1) had more than two copies of the GSTM1 or GSTT1 genes.
The frequency of GSTM1 and GSTT1 null genotypes varies in different populations. Consequently, GSTM1 null genotype was more frequent in Asians (0.23 to 0.66),16,18 Caucasians (0.45 to 0.57),16,19 and Arabs (0.50 to 0.63)20,21 compared to Africans (0.11 to 0.44).19 On the other hand, GSTT1 null genotype was shown to be more frequent among Asians (0.35 to 0.52),16,18 Africans (0.20 to 0.47)19 and Arabs (0.20 to 0.37)20,21 in contrast to Caucasians (0.14 to 0.28).16,19 The highest frequency of double GSTM1 and GSTT1 null genotypes was observed in Asians (0.27),22 followed by Arabs (0.21),21 and Africans (0.15).23 Furthermore, evidence for GSTM1 or GSTT1 gene duplication has been identified at a significantly higher frequency in a study involving a Caribbean population descended from Africa (3.4%) than Caucasians (0.2%).24,25 Indeed, our findings on the frequency of individual and combined (double) null genotypes and the percentage of more than two copies of the genes are consistent with previous studies involving Africans and populations descended from Africa.
In our study, GSTT1 carriers had a significantly lower risk of liver cirrhosis than individuals with null genotypes, and specifically, patients with one copy of the GSTT1 gene had a significantly lower risk of liver cirrhosis compared with null genotypes. Furthermore, patients with ≥2 combined GSTM1 and GSTT1 copies had a significantly lower risk of liver cirrhosis than those with a dual null genotype. In this regard, no observational studies exploring the relation between GSTT1 and GSTM1 copy numbers and the risk of liver cirrhosis are available for comparison. However, several studies have reported the association between GSTM1 and GSTT1 polymorphisms and the risk of liver disease, including drug-induced liver injury,6,26 alcoholic cirrhosis,27 viral and non-viral HCC,28 non-alcoholic fatty liver disease,29 and HCV associated cirrhosis.30 However, all previous studies reported the association based on comparing carriers (irrespective of copy number of the genes) and null genotypes of GSTM1 and GSTT1. For instance, a recent meta-analysis reported an increased risk of HCC in patients with GSTM1 or GSTT1 null genotypes, and double null genotypes of GSTM1 and GSTT1.28
Our study also demonstrated a significant gene dose relationship between the sum of GSTM1 and GSTT1 copies and the risk of liver cirrhosis. This could support the notion that the activity of enzymes encoded by GSTM1 and GSTT1 genes is directly proportional to their copy numbers.31,32 Thus, having more copy numbers could be protective against oxidative stress and the associated inflammation due to various insults to the hepatocytes, including viral and non-viral agents.
We further investigated whether GSTM1 and GSTT1 genotypes indirectly contribute to the levels of clinical chemistry tests. Accordingly, ≥2 copies of combined GSTM1 and GSTT1 genes were associated with a significant reduction in ALT and AST levels, respectively. Moreover, a significant decrease in AST level was observed in GSTT1 carriers compared with null genotypes. To the best of our knowledge, there are no studies focusing on the impact of GSTM1 and GSTT1 CNVs on clinical chemistry parameters of liver cirrhosis patients. There are, however, few studies that attempted to investigate the contribution of GSTM1 and GSTT1 to the liver biochemistry tests. For example, a study conducted among HCV-infected Brazilian cirrhosis and HCC patients reported no significant difference between carriers and null genotypes of these genes in terms of the blood level of clinical chemistry parameters.30 By contrast, our study demonstrated significant difference in AST levels between GSTT1 carriers and null genotypes of liver cirrhosis patients apart from the observed differences in ≥2 copies of combined GSTM1 and GSTT1 mentioned above. This discrepancy might arise from the fact that the mentioned study compared only null vs carriers of the genes but our study further investigated the copy numbers in carriers of the genes. Besides, differences in race and etiology of liver cirrhosis could also explain the discrepancies. Exposure to HBV and AFB1 are the most important etiologies of HCC and liver cirrhosis in Africa and Asia, whereas HCV infection and alcohol consumption are the main drivers in America, Oceania, and Europe.33–35 Indeed, in our recent study, HBV infection and AFB1 exposure were the major culprits of liver cirrhosis in Eastern Ethiopia.9
On the other hand, few studies have reported the significance of null genotypes of the GSTM1 and GSTT1 genes for changes in the serum levels of ALT and AST due to exposure to hepatotoxic drugs, which have reactive free radicals as intermediate metabolites.6,36,37 For instance, a study done among Tunisian epileptic patients treated with carbamazepine, known to have 10, 11-epoxy carbamazepine as a reactive metabolite, reported the association between GSTM1 null genotype and elevation in ALT and AST, whereas AST elevation was associated with GSTT1 null genotype.6 Thus, functional GSTM1 and GSTT1 genes might indirectly contribute to the level of AST and ALT, at least by minimizing oxidative stress and associated damage due to reactive oxygen species produced as a result of drugs and environmental toxins, including AFB1 in the hepatocytes. However, additional studies with larger sample size and inclusion of controls with a risk factor would be required for substantiating these observations.
Conclusions
Our findings suggest that a single copy number of GSTT1, and ≥2 copies of combined GSTM1 and GSTT1 are associated with reduced risk of liver cirrhosis in Ethiopians. Besides, ≥2 copies of GSTM1 and GSTT1 genes were associated with a substantial decrease in ALT and AST levels, respectively. These findings highlight the significance of gene–environment interactions in the multifactorial development of liver cirrhosis.
Acknowledgments
We would like to express our gratitude to the study participants and staff members of HFCSUH for their support during data collection. The authors would also like to acknowledge Addis Ababa University for the financial support to carry out the study. Moreover, the authors would like to thank Armauer Hansen Research Institute for laboratory access to determine GSTM1 and GSTT1 CNV and the British Council Partnership grant on food safety to the University of Leeds, School of Food Science and Nutrition for providing the kits for genetic analysis. Special thanks also goes to Hang Wu (University of Leeds) for her immense technical support.
Author Contributions
All authors made a significant contribution to the work reported whether that is in the conception, study design, execution, acquisition of data, analysis, and interpretation, or in all these areas; took part in drafting, revising, or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that there are no conflicts of interest in this work.
References
1. Mazari AMA, Zhang L, Ye Z-W, et al. The multifaceted role of glutathione s-transferases in health and disease. Biomolecules. 2023;13(4):688. doi:10.3390/biom13040688
2. Ge B, Song Y, Zhang Y, et al. Glutathione S-transferase M1 (GSTM1) and T1 (GSTT1) null polymorphisms and the risk of hypertension: a meta-analysis. PLoS One. 2015;10(3):e0118897. doi:10.1371/journal.pone.0118897
3. Qu K, Liu SS, Wang ZX, et al. Polymorphisms of glutathione S-transferase genes and survival of resected hepatocellular carcinoma patients. World J Gastroenterol. 2015;21(14):4310–4322. doi:10.3748/wjg.v21.i14.4310
4. Ginsberg G, Smolenski S, Hattis D, et al. Genetic Polymorphism in Glutathione Transferases (GST): population distribution of GSTM1, T1, and P1 conjugating activity. J Toxicol Environ Health B Crit Rev. 2009;12(5–6):389–439. doi:10.1080/10937400903158375
5. Chaudhary P, Janmeda P, Docea AO, et al. Oxidative stress, free radicals and antioxidants: potential crosstalk in the pathophysiology of human diseases. Review. Front Chem. 2023;11:1158198.
6. Chbili C, Hassine A, Fathallah N, et al. Glutathione S-transferase M1 and T1 polymorphisms and the risk of mild hepatotoxicity induced by carbamazepine in a Tunisian population study. BMC Neurol. 2018;18(1):24. doi:10.1186/s12883-018-1013-8
7. Cichoż-Lach H, Michalak A. Oxidative stress as a crucial factor in liver diseases. World J Gastroenterol. 2014;20(25):8082–8091. doi:10.3748/wjg.v20.i25.8082
8. Kapahtia S, Hazam RK, Asim M, et al. Role of glutathione S transferase M1 and T1 gene polymorphism in hepatitis B related liver diseases and cryptogenic cirrhosis. J Clin Exp Hepatol. 2018;8(2):169–172. doi:10.1016/j.jceh.2017.05.208
9. Mekuria A, Xia L, Ahmed TA, et al. Contribution of aflatoxin B1 exposure to liver cirrhosis in Eastern Ethiopia: a case-control study. Int J Gen Med. 2023;16:3543–3553. doi:10.2147/IJGM.S425992
10. Cao W, Yu P, Yang K, et al. Aflatoxin B1: metabolism, toxicology, and its involvement in oxidative stress and cancer development. Toxicol Mech Methods. 2022;32(6):395–419. doi:10.1080/15376516.2021.2021339
11. Dohnal V, Wu Q, Kuča K. Metabolism of aflatoxins: key enzymes and interindividual as well as interspecies differences. Arch Toxicol. 2014;88(9):1635–1644. doi:10.1007/s00204-014-1312-9
12. Sun CA, Wang LY, Chen CJ, et al. Genetic polymorphisms of glutathione S-transferases M1 and T1 associated with susceptibility to aflatoxin-related hepatocarcinogenesis among chronic hepatitis B carriers: a nested case-control study in Taiwan. Carcinogenesis. 2001;22(8):1289–1294. doi:10.1093/carcin/22.8.1289
13. Long XD, Ma Y, Wei YP, et al. Study on the detoxication gene GSTM1-GSTT1-null and susceptibility to aflatoxin B1 related hepatocellular carcinoma in Guangxi. Zhonghua Liu Xing Bing Xue Za Zhi. 2005;26(10):777–781.
14. Zhang Y, Meng X, Ma Z, et al. Association of androgen-receptor gene mutations with the copy number of androgen-receptor silk protein A complex and Glutathione-S-Transferases T1 and M1 in prostate cancer patients. Genet Res. 2023;2023:5956951. doi:10.1155/2023/5956951
15. Mawick A, Pfeiffer H, Vennemann M. Sudden infant death syndrome: deletions of glutathione-S-transferase genes M1 and T1 and tobacco smoke exposure. Int J Legal Med. 2021;135(4):1375–1383. doi:10.1007/s00414-021-02556-5
16. Nakanishi G, Pita-Oliveira M, Bertagnolli LS, et al. Worldwide systematic review of GSTM1 and GSTT1 null genotypes by continent, ethnicity, and therapeutic area. Omics. 2022;26(10):528–541. doi:10.1089/omi.2022.0090
17. Nugrahaningsih DAA, Wihadmadyatami H, Widyarini S, et al. A review of the GSTM1 null genotype modifies the association between air pollutant exposure and health problems. Int J Genomics. 2023;2023:4961487. doi:10.1155/2023/4961487
18. Alshagga MA, Mohamed N, Nazrun SA, et al. Frequencies of glutathione s-transferase (GSTM1, GSTM3 and GSTT1) polymorphisms in a Malaysian population. Arch Med Sci. 2011;7(4):572–578. doi:10.5114/aoms.2011.24123
19. Piacentini S, Polimanti R, Porreca F, et al. GSTT1 and GSTM1 gene polymorphisms in European and African populations. Mol Biol Rep. 2011;38(2):1225–1230. doi:10.1007/s11033-010-0221-0
20. Mansour AA, Saleh OM, Askar T, et al. Frequency of glutathione-S-transferase null-M1 and null-T1 genotypes among the Turabah population in Saudi Arabia. Genet Mol Res. 2015;14(4):16863–16871. doi:10.4238/2015.December.14.13
21. Salem AH, Yaqoob A, Ali M, et al. Genetic polymorphism of the glutathione S-transferase M1 and T1 genes in three distinct Arab populations. Dis Markers. 2011;31(5):311–316. doi:10.1155/2011/796520
22. Guo X, O’Brien SJ, Zeng Y, et al. GSTM1 and GSTT1 gene deletions and the risk for nasopharyngeal carcinoma in Han Chinese. Cancer Epidemiol Biomarkers Prev. 2008;17(7):1760–1763. doi:10.1158/1055-9965.EPI-08-0149
23. Kiendrebeogo IT, Zoure AA, Sorgho PA, et al. Glutathione S-transferase M1 (GSTM1) and T1 (GSTT1) variants and breast cancer risk in Burkina Faso. Biomol Concepts. 2019;10(1):175–183. doi:10.1515/bmc-2019-0020
24. Nørskov M, Frikke-Schmidt R, Bojesen S, et al. Copy number variation in glutathione-S-transferase T1 and M1 predicts incidence and 5-year survival from prostate and bladder cancer, and incidence of corpus uteri cancer in the general population. Pharmacogenomics J. 2011;11(4):292–299. doi:10.1038/tpj.2010.38
25. Emeville E, Broquère C, Brureau L, et al. Copy number variation of GSTT1 and GSTM1 and the risk of prostate cancer in a Caribbean population of African descent. PLoS One. 2014;9(9):e107275. doi:10.1371/journal.pone.0107275
26. Li C, Long J, Hu X, et al. GSTM1 and GSTT1 genetic polymorphisms and risk of anti-tuberculosis drug-induced hepatotoxicity: an updated meta-analysis. Eur J Clin Microbiol Infect Dis. 2013;32(7):859–868. doi:10.1007/s10096-013-1831-y
27. Ladero JM, Martínez C, García-Martin E, et al. Polymorphisms of the glutathione S-transferases mu-1 (GSTM1) and theta-1 (GSTT1) and the risk of advanced alcoholic liver disease. Scand J Gastroenterol. 2005;40(3):348–353. doi:10.1080/00365520510012109
28. Li S, Xue F, Zheng Y, et al. GSTM1 and GSTT1 null genotype increase the risk of hepatocellular carcinoma: evidence based on 46 studies. Cancer Cell Int. 2019;19(1):76. doi:10.1186/s12935-019-0792-3
29. Damavandi N, Zeinali S. Association of xenobiotic-metabolizing enzymes (GSTM1 and GSTT 1), and pro-inflammatory cytokines (TNF-α and IL-6) genetic polymorphisms with non-alcoholic fatty liver disease. Mol Biol Rep. 2021;48(2):1225–1231. doi:10.1007/s11033-021-06142-1
30. Araujo OC, de Paula VS, Do Ó KM, et al. Association of Polymorphisms in the Glutathione S-Transferase Theta-1 gene with cirrhosis and hepatocellular carcinoma in Brazilian patients with chronic hepatitis C. Vaccines. 2021;9(8):831. doi:10.3390/vaccines9080831
31. Sprenger R, Schlagenhaufer R, Kerb R, et al. Characterization of the glutathione S-transferase GSTT1 deletion: discrimination of all genotypes by polymerase chain reaction indicates a trimodular genotype-phenotype correlation. Pharmacogenetics. 2000;10(6):557–565. doi:10.1097/00008571-200008000-00009
32. Girault I, Lidereau R, Bièche I. Trimodal GSTT1 and GSTM1 genotyping assay by real-time PCR. Int J Biol Markers. 2005;20(2):81–86. doi:10.1177/172460080502000201
33. Fassio E, Díaz S, Santa C, et al. Etiology of hepatocellular carcinoma in Latin America: a prospective, multicenter, international study. Ann Hepatol. 2010;9(1):63–69. doi:10.1016/S1665-2681(19)31681-3
34. El-Kassas M, Elbadry M. Hepatocellular carcinoma in Africa: challenges and opportunities. Front Med. 2022;9:899420. doi:10.3389/fmed.2022.899420
35. Díaz LA, Ayares G, Arnold J, et al. Liver Diseases in Latin America: current status, unmet Needs, and opportunities for improvement. Curr Treat Options Gastroenterol. 2022;20(3):261–278. doi:10.1007/s11938-022-00382-1
36. Roy B, Chowdhury A, Kundu S, et al. Increased risk of antituberculosis drug-induced hepatotoxicity in individuals with glutathione S-transferase M1 ‘null’ mutation. J Gastroenterol Hepatol. 2001;16(9):1033–1037.
37. Watanabe I, Tomita A, Shimizu M, et al. A study to survey susceptible genetic factors responsible for troglitazone-associated hepatotoxicity in Japanese patients with type 2 diabetes mellitus. Clin Pharmacol Ther. 2003;73(5):435–455.
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.