Back to Journals » Drug Design, Development and Therapy » Volume 9
Apolipoprotein M regulates the orphan nuclear receptor LRH-1 gene expression through binding to its promoter region in HepG2 cells
Authors Pan Y, Zhou H, Zhou H, Hu M, Tang L
Received 2 December 2014
Accepted for publication 26 February 2015
Published 24 April 2015 Volume 2015:9 Pages 2375—2382
DOI https://doi.org/10.2147/DDDT.S78496
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Professor Shu-Feng Zhou
Yi Pan,1,2,* Hou-gang Zhou,1,* Hui Zhou,3 Min Hu,1 Li-jun Tang2
1Clinical Laboratory, The Second Xiangya Hospital of Central South University, Changsha, Hunan, People’s Republic of China; 2Molecular Biology Research Center, School of Life Sciences, Central South University, Changsha, Hunan, People’s Republic of China; 3Tumor Hospital of Xiangya School of Medicine, Central South University, Changsha, Hunan, People’s Republic of China
*These authors contributed equally to this work
Abstract: Apolipoprotein M (ApoM) is predominantly located in the high-density lipoprotein in human plasma. It has been demonstrated that ApoM expression could be regulated by several crucial nuclear receptors that are involved in the bile acid metabolism. In the present study, by combining gene-silencing experiments, overexpression studies, and chromatin immunoprecipitation assays, we showed that ApoM positively regulated liver receptor homolog-1 (LRH-1) gene expression via direct binding to an LRH-1 promoter region (nucleotides -406/-197). In addition, we investigated the effects of farnesoid X receptor agonist GW4064 on hepatic ApoM expression in vitro. In HepG2 cell cultures, both mRNA and protein levels of ApoM and LRH-1 were decreased in a time-dependent manner in the presence of 1 µM GW4064, and the inhibition effect was gradually attenuated after 24 hours. In conclusion, our findings present supportive evidence that ApoM is a regulator of human LRH-1 transcription, and further reveal the importance of ApoM as a critical regulator of bile acids metabolism.
Keywords: bile acids, chromatin immunoprecipitation assay, farnesoid X receptor, GW4064, high-density lipoprotein
Introduction
Bile acid, the end product of cholesterol catabolism, has only been simply understood as a major pathway by which cholesterol is excreted from the body in the past decades.1 However, in recent years, increasing studies have figured out that bile acids are also important signaling molecules, not only of their own synthesis but also of energy metabolism, glycometabolism, lipid metabolism, and liver regeneration.2–5
Nuclear receptor is considered to be one of the most important regulating factors in the process of bile acids metabolism. The nuclear receptor superfamily comprises a large group of ligand-activated transcription factors that play a key regulatory role in the cellular communication of multicellular organisms.6 They are usually responsible for maintaining balance of target gene expression and regulating gene networks involved in body metabolism, development, cellular differentiation, and reproductive function, through interacting with corresponding ligands and coactivators.7,8 Nuclear receptors are categorized into three subclasses (nonsteroid hormone receptors, steroid hormone receptors, and orphan nuclear receptors) according to the type of their ligands. As two important members of the nuclear receptor superfamily, liver receptor homolog-1 (LRH-1) and farnesoid X receptor (FXR) play a pivotal role in the bile acid metabolism.9
To date, LRH-1 still remains an orphan member of the nuclear receptor superfamily, whose physiological ligands have not yet been identified, while FXR can be activated by physiological concentrations of bile acids.10,11 It has been demonstrated that activated FXR induces expression of small heterodimer partner (SHP), which represses transcription of CYP7A1, the rate-limiting enzyme in bile acid synthesis pathway, by forming a inhibition complexity with LRH-1.12 This bile acid-activated regulatory cascade has an important implication in maintenance of bile acid and cholesterol homeostasis. Since activated FXR exhibits attractive physiological functions, there is an increasing need for new FXR agonists with high binding affinity and efficacy. The most widely used FXR agonists reported to date are chenodeoxycholic acid (CDCA) as well as GW4064 and its derivatives.13,14 CDCA is the most active natural ligand for FXR, while the synthetic FXR-specific ligand GW4064 exhibits a higher efficacy and selectivity over CDCA.
Apolipoprotein M (ApoM), one of the new members of the apolipoprotein family, may have potential antiatherosclerotic properties, which may be mediated by the enhancement of reversed cholesterol transportation and/or hepatic cholesterol catabolism.15–17 Although there is no direct evidence to suggest that ApoM regulates bile acid metabolism, previous studies have demonstrated that the expression of ApoM could be regulated by most of the nuclear receptors involved in bile acid metabolism,12,18,19 and these indicated that ApoM might play a role in the process of bile acid metabolism. In the present study, we showed by combining gene-silencing experiments, overexpression studies, and chromatin immunoprecipitation (ChIP) assays that ApoM positively regulated LRH-1 gene expression via direct binding to a sequence located in the LRH-1 promoter region. Meanwhile, we investigated the effects of FXR activated by its agonist GW4064 on hepatic ApoM expression in vitro. Our findings further illustrated the importance of ApoM as a critical regulator of bile acids metabolism.
Materials and methods
Reagents
Dulbecco’s Modified Eagle’s Medium (DMEM) high glucose, Opti-MEM I Reduced Serum Medium, and fetal bovine serum were from Gibco (Carlsbad, CA, USA). Phusion High-Fidelity PCR Kit, restriction enzymes, and T4 DNA ligase were purchased from New England Biolabs (Beverly, MA, USA). Wizard SV Gel and PCR Clean-Up System was purchased from Promega Corporation (Madison, WI, USA). Escherichia coli DH5α Competent Cells were purchased from Takara (Dalian, People’s Republic of China). High Pure Plasmid Isolation Kit was purchased from Hoffman-La Roche Ltd. (Basel, Switzerland). Lipofectamine™ 2000 Transfection Reagent was purchased from Thermo Fisher Scientific (Waltham, MA, USA). RNeasy Mini Kit was purchased from Qiagen NV (Venlo, the Netherlands). High Capacity cDNA Reverse Transcription Kits and SYBR Select Master Mix were purchased from Thermo Fisher Scientific. Alexa Fluor 488 Goat Anti-Mouse and Anti-Rabbit IgG(H+L) were purchased from Molecular Probes (Rockland, ME, USA). Rabbit anti-human ApoM polyclonal antibody and mouse anti-human NR5A2/LRH-1 monoclonal antibody were purchased from Abcam (Cambridge, MA, United States). BCA Protein Assay Kit was purchased from Pierce (Rockford, IL, USA). Human kidney cDNA library was from Open Biosystems (Huntsville, AL, USA). GW4064 was obtained from Sigma-Aldrich Co. (St Louis, MO, USA).
Cell culture
Human hepatoma HepG2 cells stored by our laboratory were cultured in high glucose DMEM supplemented with 10% fetal bovine serum, 100 units/mL penicillin, and 100 μg/mL streptomycin (Thermo Fisher Scientific) at 37°C in a 5% CO2 atmosphere, and the medium was changed every 2 days. Cells were seeded in six-well cell culture plates, and grown to 50%–70% confluence. Prior to experiments, cells were washed twice with phosphate buffered saline (PBS) and serum-free DMEM without antibiotics. Then, GW4064 dissolved in dimethyl sulfoxide was added to a final concentration of 1 μM in media allowing for treatment for the indicated time.
Plasmid construction
The full sequence of human ApoM gene (NCBI Reference Sequence: NM_019101.2) was generated by polymerase chain reaction (PCR) with the forward primer 5′-CCGCTCGAG-ATGTTCCACCAAATTTGGGCAGCT-3′ (Xho I) and the reverse primer 5′-CGCGGATCCCGGTTATTGGACAGCTCACAGGCCTC-3′ (BamH I) from the human kidney cDNA library. Amplification was performed in 50 μL containing RNase free double-distilled H2O (34 μL), 5× Phusion HF buffer (10 μL), 10 mmol/L deoxynucleotide (1 μL), human kidney cDNA library (1 μL), 10 μmol/L primers (2.5 μL each), and Phusion DNA polymerase (0.5 μL). Following one denaturation step (1 minute at 98°C), 35 cycles of amplification (20 seconds at 94°C, 20 seconds at 50°C, 1 minute at 72°C) and a final elongation step of 10 minutes at 72°C were carried out. The PCR product (564 bp) was purified with Wizard SV Gel and PCR Clean-Up System and then inserted as an Xho I/BamH I fragment into the pEGFP-N1 expression vector yielding pEGFP-N1-ApoM.
siRNA-mediated gene silence
High-purity Mission siRNA-ApoM and negative control oligos were synthesized by Sigma-Aldrich Co. The Mission siRNA-ApoM comprised two reverse completed oligos (sense 5′-CUGUGGACAACAUUGUCUUdTdT-3′ and antisense 5′-AAGACAAUGUUGUCCACAGdTdT-3′) which were designed against the 267–285 bp target site of the ApoM mRNA, while the negative control oligos (sense 5′-UUCUCCGAACGUGUCACGUdTdT-3′ and antisense 5′-ACGUGACACGUUCGGAGAAdTdT-3′) lacked significant sequence homology to human ApoM mRNA or any other genes in the genome. siRNA transfection was performed using the Lipofectamine 2000 reagent and gene-silencing efficiency was evaluated by quantitative real-time (qRT)-PCR and western blotting using rabbit anti-human ApoM polyclonal antibody.
Cell transfection
Transient transfections of cells with pEGFP-N1-ApoM and Mission siRNA-ApoM were performed with the Lipofectamine 2000 reagent according to the manufacturer’s instructions. Briefly, HepG2 cells were seeded in six-well cell culture plates, and grown to 70%–90% confluence. Prior to transfection, the supernatants were removed and replaced with fresh media. Then, 4 μg pEGFP-N1-ApoM or 100 pmol Mission siRNA-ApoM were diluted into 250 μL Opti-MEM I Reduced Serum Medium. Meanwhile, for each well of cells, appropriate volumes of Lipofectamine 2000 were diluted into 250 μL Opti-MEM I Reduced Serum Medium, and incubated at room temperature for 5 minutes. After that, these two dilutions were combined, mixed gently, and incubated for 20 minutes at room temperature to allow for DNA–Lipofectamine 2000 complexes to form. The DNA–Lipofectamine 2000 complexes (500 μL) were added directly to each well containing cells and mixed gently. Twenty-four hours post transfection, cells were collected and assayed for transfection efficiency.
qRT-PCR
Total RNA was extracted using the RNeasy Mini Kit according to the manufacturer’s instructions. One microgram of this RNA was reverse transcribed with RT random primers using a High Capacity cDNA Reverse Transcription Kit, and the cDNAs produced were used for qRT-PCR amplification with the primers shown in Table 1. qRT-PCR amplification, detection, and analysis was performed on a PRISM 7500 Real-Time PCR System (Thermo Fisher Scientific) using SYBR Select Master Mix (Thermo Fisher Scientific) following the manufacturer’s protocol. Transcript generated from the GAPDH gene was used as an internal control for normalization.
  | Table 1 Primers used in qRT-PCR |
Western blotting
For the extraction of total protein, cell-culture medium was completely aspirated from the six-well culture plates, and cells were washed twice with pre-chilled PBS and lysed with 250 μL RIPA solution. Then, the lysate was collected with a rubber policeman, transferred into a pre-cold Eppendorf (EP) tube, mixed gently, and incubated for 30 minutes at 4°C. After that, the EP tube was centrifuged at 16,000× g for 20 minutes at 4°C, and finally the supernatant was transferred into a new pre-cold EP tube. The extracted total protein sample was quantified using a BCA Protein Assay Kit according to the manufacturer’s protocol.
Thereafter, 20 μg total protein per lane was added in the same volume of 2× loading buffer and subjected to denaturation at 100°C for 5 minutes, then electrophoresed on 15% sodium dodecyl sulfate polyacrylamide gel electrophoresis at 100 mA for 3 hours, and finally transferred onto polyvinylidene fluoride membrane using a Mini Trans-Blot Cell (Bio-Rad Laboratories Inc., Hercules, CA, USA). Anti-ApoM, anti-LRH-1, and anti-GAPDH antibodies were used as primary antibodies. After incubation with a, signals were detected using an ECL Advanced Western blot analysis system (Amersham Pharmacia Biotech Inc., Piscataway, NJ, USA).
Flow cytometry
The method for examination of cell surface or intracellular antigen in fixed cells by flow cytometry was modified based on the indirect immunofluorescent labeling techniques reported by Cunningham.20 Briefly, cells harvested and washed twice with PBS. All cells were incubated in the dark for 30 min on ice primary antibodies respectively. After washing twice with PBS, Alexa Fluor 488 conjugated antibody was added and incubated 30 min on ice. Cells were pelleted and resuspended in 500 μL PBS. The results were analyzed with FlowJo software.
ChIP assays
ChIP assays were performed using the EZ ChIP KIT (MD Millipore, Billerica, MA, USA) following the manufacturer’s instructions. Examination of ApoM recruitment to human LRH-1 promoter was performed using ChIP with chromatin from HepG2 cells and ApoM polyclonal antibodies (Abcam) or IgGs as isotype control (Santa Cruz Biotechnology Inc., Dallas, TX, USA). Human LRH-1 promoter occupancy was then assessed by PCR amplification of its regions using primers shown in Table 2. A pair of primers (ApoM-49F: TCATTAGCAGGTGAAAGGGTCAAGG; ApoM-42R: GCCTTAACTGCTCTCTCCCCTACTG) designed according to a fragment of ApoM gene was used as a control. PCR products were analyzed by electrophoresis on a 1.5% agarose gel.
  | Table 2 Primers used in chromatin immunoprecipitation assays |
Statistical analysis
Results are shown as mean ± standard deviation. Analysis of variance was performed with SPSS 15.0 software for Windows using one-way analysis of variance and pair-wise comparison with t-test. P<0.05 was considered statistically significant.
Results
Modulation of ApoM expression by transfection with pEGFP-N1-ApoM or ApoM siRNA in HepG2 cells
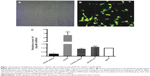
An increasing number of studies have implied that ApoM may play a critical role in the process of bile acid metabolism. It has been demonstrated that ApoM expression can be regulated by some nuclear receptors including LRH-1. To elucidate the role of ApoM in the interaction with LRH-1 in vitro, gene overexpression and gene silencing experiments were employed in the liver-derived cell line, HepG2 cells. Firstly, an ApoM overexpression plasmid pEGFP-N1-ApoM, in which the ApoM was located upstream of an EGFP gene and formed an ApoM-EGFP fusion protein, was constructed. Thereafter, the plasmid pEGFP-N1-ApoM and Mission siRNA-ApoM along with their controls, pEGFP-N1 and negative control oligos, were transiently transfected into HepG2 cells, respectively. Twenty-four hours post-transfection, bright green fluorescent signals were detected in pEGFP-N1-ApoM-transfected HepG2 cells, and the transfection efficiency was estimated beyond 80% (Figure 1A and B). Meanwhile, pEGFP-N1-ApoM infection in HepG2 cells resulted in a sharp increase in ApoM mRNA levels compared with control pEGFP-N1-infected cells, while in Mission siRNA-ApoM-transfected cells, the ApoM mRNA level was decreased significantly compared with control siRNA-infected cells (Figure 1C).
Effects of upregulation or inhibition of ApoM expression on LRH-1 in HepG2 cells
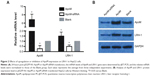
In order to investigate the effects of ApoM on LRH-1 gene expression, the mRNA and protein expression level of LRH-1 in infected cells were examined. As shown in Figure 2A, decreased ApoM mRNA expression level in Mission siRNA-ApoM-transfected HepG2 cells was associated with a 50% reduction in LRH-1 mRNA level compared with control, and upregulated ApoM mRNA expression in pEGFP-N1-ApoM-transfected cells led to a 50% increase of LRH-1 mRNA level. In agreement with these findings, it was shown by Western blotting that LRH-1 expression was also significantly decreased in response to ApoM siRNA interference, while in ApoM overexpression cells, the LRH-1 protein was accordingly increased (Figure 2B). In summary, the above findings strongly suggest that ApoM positively regulated LRH-1 expression at both transcription and protein expression levels in hepatic cells.
ApoM binds to the promoter region of human LRH-1 gene
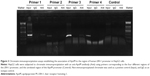
To clarify the molecular mechanism by which ApoM regulates LRH-1 expression, a ChIP assay that examined association of ApoM to the human LRH-I gene promoter was performed. As shown in Figure 3, an antibody against ApoM could efficiently bind and immunoprecipitate the −424/−83 region of LRH-1 promoter but not the −196/+65 region of the LRH-1 promoter. But there was also a weak band in the ChIP sample with antibody for primer 3. In addition, the input band is too dim for primer 2, so the −692/−407 and −875/−676 sequences might also be the promoter binding regions. And a more precise analysis will be conducted in future experiments. Taken together, these results suggested that the −406/−197 region of the LRH-1 promoter was probably the ApoM binding site. In control reactions, ApoM was not found to bind to its own gene sequence, which confirmed that the binding of ApoM is specific.
Effects of GW4064 treatment on LRH-1 mRNA and protein expression in HepG2 cells
Because a previous study has reported that the synthetic FXR agonist GW4064 suppressed ApoM expression via a FXR-SHP-LRH-1 cascade in primary human hepatocytes,12 we attempted to investigate the effects of GW4064 on ApoM and LRH-1 expression in HepG2 cells. After treatment with 1 μM GW4064, HepG2 cells were harvested at 0, 6, 12, 24, 36, 48, and 60 hours and subjected to qRT-PCR, Western blotting, and flow cytometry examination of ApoM and LRH-1 expression changes, respectively. Results showed that 1 μM GW4064 decreased expression of ApoM and LRH-1 at both the mRNA level and protein level in a time-dependent manner before the time-point of 24 hours (Figure 4A–C). While the inhibition effect was attenuated after 24 hours, the expression level of ApoM and LRH-1 were gradually restored as indicated by Western blotting analysis.
Discussion
ApoM, a novel member of the apolipoprotein family, is located predominantly in high-density lipoprotein in human plasma.21,22 Up to now, the biological functions of ApoM have not been well understood, and it is speculated that ApoM may be involved in reverse cholesterol transport as well as antiatherosclerotic and antioxidant effects based on its location. Previous studies have reported that ApoM could be regulated by many factors mainly including nuclear receptors and cytokines, most of which act as critical factors in bile acid metabolism.12,18,19 It indicates that ApoM may play vital a role in the process of bile acid metabolism. However, the molecular mechanism that ApoM involves in regulating the bile acid metabolism process remains to be clarified.
In this study, we showed for the first time that ApoM directly regulates LRH-1 gene expression which is believed to be a key player in bile acid and cholesterol metabolism. The orphan nuclear receptor LRH-1 is mainly expressed in the liver, pancreas, and intestine,23 and is known to play a pivotal role in the transcriptional regulation of CYP7A1 and CYP7B1, the rate-limiting enzymes for the classic and alternative bile acid biosynthetic pathways, respectively.24 ApoM is also one of the target genes transcriptionally regulated by LRH-1. Venteclef et al12 has established that LRH-1 positively regulates human ApoM transcription by directly binding to a 5′-CAAGG-3′ motif present in the proximal ApoM promoter region. Our data show that when ApoM expression is upregulated or decreased by transfection, the LRH-1 expression also changes following the same trend in response. These results strongly suggest that LRH-1 may be an ApoM target gene in HepG2 cells. Furthermore, a ChIP assay confirmed our speculation that ApoM directly regulated human LRH-1 transcription by association to its promoter region. These findings indicate that ApoM and LRH-1 may play a critical role in bile acid metabolism through interaction with each other.
Meanwhile, it has been reported that ApoM gene expression is subjected to regulation by agonists of FXR in vivo and in vitro. Venteclef et al12 showed that treatment of primary hepatocytes with the FXR ligand GW4064 (1 μM) or CDCA (50 μM) suppressed ApoM expression via a FXR-SHP-LRH-1 cascade. Another study by Zhang et al19 found that T0901317, the dual liver X receptor/FXR agonist, downregulated ApoM gene expression in both mouse liver and HepG2 cells. Subsequently, the study by Venteclef et al12 verified that inhibition of ApoM gene transcription by T0901317 could be due to the activation of the FXR/SHP pathway that inhibits LRH-1-mediated transactivation of ApoM in hepatic cells. In line with these findings, we found that treatment of HepG2 cells with 1 μM FXR agonist GW4064 resulted in a significant decrease of both mRNA and protein expression level of ApoM in a time-dependent manner before 24 hours, while after treatment of GW4064 for 24 hours, ApoM expression increased gradually and was kept at a relative stable level finally after 48 hours. Reasons should be attributed to the negative feedback regulation of bile acid metabolism. It is very interesting to find that 1 μM GW4064 also resulted in LRH-1 expression changing in the same manner, which could possibly be explained by the positive regulation of LRH-1 expression by ApoM. Based on the above results, it is tempting to speculate that ApoM could act as a key player in bile acid metabolism through direct interaction with LRH-1 in the FXR-SHP-LRH-1 pathway.
In conclusion, we have presented supportive evidence for the first time that ApoM, a recently discovered high-density lipoprotein apolipoprotein, is a regulator of human LRH-1 transcription. These new findings contribute to a better understanding of the molecular basis of the bile acid metabolism, and may help to explore a new way to clarify the pathogenesis of bile acid metabolism systematic disease, as well as providing effective early diagnosis and interventional therapy.
Acknowledgments
This work was supported by Hunan Provincial Natural Science Fund (2013J5043), Changsha Science and Technology (K1207031-31) and Science and Technology Innovation Investment Programs of Development and Reform Commission of Hunan Province([2014] No 658 document, the 25th plan of Central South University).
Author contributions
YP, HGZ, MH, and LJT conceived and designed the research. YP, HGZ, and HZ performed all experiments. MH and LJT analyzed the data and were responsible for the work. All authors contributed toward data analysis, drafting and revising the paper and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
Lu TT, Makishima M, Repa JJ, et al. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol Cell. 2000;6:507–515. | ||
Kobayashi M, Ikegami H, Fujisawa T, et al. Prevention and treatment of obesity, insulin resistance, and diabetes by bile acid-binding resin. Diabetes. 2007;56:239–247. | ||
Zhang Y, Lee FY, Barrera G, et al. Activation of the nuclear receptor FXR improves hyperglycemia and hyperlipidemia in diabetic mice. Proc Natl Acad Sci U S A. 2006;103:1006–1011. | ||
Watanabe M, Houten SM, Mataki C, et al. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature. 2006;439:484–489. | ||
Inagaki T, Choi M, Moschetta A, et al. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2005;2:217–225. | ||
Clarke ND, Berg JM. Zinc fingers in Caenorhabditis elegans: finding families and probing pathways. Science. 1998;282:2018–2022. | ||
Schulman IG. Nuclear receptors as drug targets for metabolic disease. Adv Drug Deliv Rev. 2010;62:1307–1315. | ||
Huang HJ, Schulman IG. Regulation of metabolism by nuclear hormone receptors. Prog Mol Biol Transl Sci. 2009;87:1–51. | ||
Repa JJ, Mangelsdorf DJ. Nuclear receptor regulation of cholesterol and bile acid metabolism. Curr Opin Biotechnol. 1999;10:557–563. | ||
Makishima M, Okamoto AY, Repa JJ, et al. Identification of a nuclear receptor for bile acids. Science. 1999;284:1362–1365. | ||
Parks DJ, Blanchard SG, Bledsoe RK, et al. Bile acids: natural ligands for an orphan nuclear receptor. Science. 1999;284:1365–1368. | ||
Venteclef N, Haroniti A, Tousaint JJ, Talianidis I, Delerive P. Regulation of anti-atherogenic apolipoprotein M gene expression by the orphan nuclear receptor LRH-1. J Biol Chem. 2008;283:3694–3701. | ||
Pellicciari R, Fiorucci S, Camaioni E, et al. 6alpha-ethyl-chenodeoxycholic acid (6-ECDCA), a potent and selective FXR agonist endowed with anticholestatic activity. J Med Chem. 2002;45:3569–3572. | ||
Maloney PR, Parks DJ, Haffner CD, et al. Identification of a chemical tool for the orphan nuclear receptor FXR. J Med Chem. 2000;43:2971–2974. | ||
Plomgaard P, Dullaart RP, de Vries R, Groen AK, Dahlbäck B, Nielsen LB. Apolipoprotein M predicts pre-beta-HDL formation: studies in type 2 diabetic and nondiabetic subjects. J Intern Med. 2009;266:258–267. | ||
Wolfrum C, Poy MN, Stoffel M. Apolipoprotein M is required for prebeta-HDL formation and cholesterol efflux to HDL and protects against atherosclerosis. Nat Med. 2005;11:418–422. | ||
Liu M, Seo J, Allegood J, et al. Hepatic apolipoprotein M (ApoM) overexpression stimulates formation of larger ApoM/sphingosine 1-phosphate-enriched plasma high density lipoprotein. J Biol Chem. 2014;289:2801–2814. | ||
Mosialou I, Zannis VI, Kardassis D. Regulation of human apolipoprotein m gene expression by orphan and ligand-dependent nuclear receptors. J Biol Chem. 2010;285:30719–30730. | ||
Zhang X, Zhu Z, Luo G, Zheng L, Nilsson-Ehle P, Xu N. Liver X receptor agonist downregulates hepatic apoM expression in vivo and in vitro. Biochem Biophys Res Commun. 2008;371:114–117. | ||
Cunningham RE. Indirect immunofluorescent labeling of fixed cells. Methods Mol Biol. 2010;588:335–339. | ||
Xu N, Dahlbäck B. A novel human apolipoprotein (apoM). J Biol Chem. 1999;274:31286–31290. | ||
Luo G, Zhang X, Nilsson-Ehle P, Xu N. Apolipoprotein M. Lipids Health Dis. 2004;3:21. | ||
Fayard E, Auwerx J, Schoonjans K. LRH-1: an orphan nuclear receptor involved in development, metabolism and steroidogenesis. Trends Cell Biol. 2004;14:250–260. | ||
Nitta M, Ku S, Brown C, Okamoto AY, Shan B. CPF: an orphan nuclear receptor that regulates liver-specific expression of the human cholesterol 7alpha-hydroxylase gene. Proc Natl Acad Sci U S A. 1999;96:6660–6665. |
 © 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.