Back to Journals » OncoTargets and Therapy » Volume 13
ZFAS1 Exerts an Oncogenic Role via Suppressing miR-647 in an m6A-Dependent Manner in Cervical Cancer
Authors Yang Z, Ma J, Han S, Li X, Guo H, Liu D
Received 29 July 2020
Accepted for publication 28 September 2020
Published 17 November 2020 Volume 2020:13 Pages 11795—11806
DOI https://doi.org/10.2147/OTT.S274492
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Federico Perche
Zhijuan Yang,1,* Jingwen Ma,1,* Shuxia Han,1 Xiaowen Li,1 Hua Guo,1 Dongtao Liu2
1Gynecology Department, General Hospital of Ningxia Medical University, Yinchuan City, Ningxia Province, People’s Republic of China; 2Gastrointestinal Department, General Hospital of Ningxia Medical University, Yinchuan City, Ningxia Province, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Dongtao Liu
Gastrointestinal Department, General Hospital of Ningxia Medical University, No. 804, Shengli South Street, Xingqing District, Yinchuan City, Ningxia Province 750004, People’s Republic of China
Email [email protected]
Background: Cervical cancer (CC) is the second serious health threat in women worldwide. LncRNA (ZNFX1 antisense RNA 1) ZFAS1 has been observed to abnormally express in human cancers. However, the expression pattern, clinical significance and molecular mechanism of ZFAS1 have not been thoroughly studied in CC.
Methods: qRT-PCR was performed to examine the differential expression of ZFAS1 in CC tissues and adjacent normal cervical tissues. Gain- and loss-of-function experiments were constructed to test the functional role of ZFAS1 in CC by CCK-8, colony formation, transwell and xenograft models assays. Luciferase reporter, RNA immunoprecipitation (RIP), methylated RNA immunoprecipitation (MeRIP), RNA pull-down assays were used to reveal the underlying mechanisms.
Results: We found that ZFAS1 was significantly upregulated in CC tissues. Elevation of ZFAS1 correlated with advanced FIGO stage, lymph node and distant metastasis, and also indicated poor overall survival in patients with CC. Functional experiments demonstrated that ZFAS1 promoted CC cell proliferation, migration and invasion in vitro, and facilitated tumor growth and metastasis in vivo. Mechanistic investigation revealed that ZAFS1 sequestered miR-647, and this RNA–RNA interaction is regulated by METLL3-mediated m6A modification.
Conclusion: Our findings elucidate the functional roles of ZFAS1 and its m6A modification in CC cells and indicate that ZFAS1 may be a promising target for CC treatment.
Keywords: m6A modification, ceRNA, growth, metastasis
Introduction
Cervical cancer (CC) is the second serious health threat in women worldwide.1 Although great strides have been made in the diagnostic and therapeutic means for CC in the past decades, the prognosis of patients with CC remains unsatisfactory because of its late detection, recurrence and distant metastasis.2 Therefore, it is urgently needed to identify the key molecule and reveal its underlying mechanism of CC occurrence and progression.
Long-noncoding RNAs (lncRNAs) are a class of noncoding RNAs, which are longer than 200 nucleotides (nts). LncRNAs are capable to regulate gene expression at epigenetic, transcriptional and posttranscriptional processes, and affect the posttranslational modification of its interacting protein.3,4 A growing number of studies has demonstrated that lncRNAs could function as either oncogenes or tumor suppressors through their diverse regulatory mechanisms.5,6 LncRNA (ZNFX1 antisense RNA 1) ZFAS1 is the antisense transcript of the gene ZNFX1 and has been observed to abnormally express in human cancers. In different types of cancers, ZFAS1 plays opposite functions. For example, ZFAS1 was overexpressed in hepatocellular carcinoma tissues and promotes malignancy via regulating miR-624.7 In ovarian cancer, ZFAS1 is highly expressed and accelerate cell proliferation, migration, invasion, and induces cisplatin resistance by directly suppressing miR-548e expression.8 ZFAS1 was found to be downregulated in breast tumors compared to normal tissue.9 However, the expression pattern, clinical significance and molecular mechanism of ZFAS1 has not been thoroughly studied in CC.
In this study, we found that upregulation of ZFAS1 is associated with advanced FIGO stage, lymph node and distant metastasis, and indicates poor prognosis in patients with CC. Functional experiments showed that ZFAS1 plays an oncogenic role to promote CC proliferation and metastasis. Mechanistically, our results demonstrated that ZFAS1 sequestered miR-647 in a m6A-dependent manner. Collectively, our research revealed the clinical significance and regulatory mechanism of ZFAS1 in CC.
Materials and Methods
Tissue Collection
CC tissues and adjacent normal cervical tissues were collected from CC patients who underwent surgical resection at the General Hospital of Ningxia Medical University between January 2013 and December 2019. None of these patients had received other treatment before surgery, including chemotherapy and radiotherapy. This study was approved by the Institutional Ethical Review Board of General Hospital of Ningxia Medical University, and written informed consent was obtained from all patients in this study.
Cell Culture
Hela, SiHa, C33A, CaSki and 293T cell lines were obtained from the Cell Bank of Type Culture Collection (Shanghai, China). All cells were cultured in DMEM medium supplemented with 10% FBS (Gibco) in an incubator containing 5% CO2 at 37°C. To construct stable Caski cells with ZFAS1 or METTL3 knockdown, the ZFAS1 shRNA (shZFAS1) or METTL3 shRNA (shMETTL3) or negative control shRNA (shNC) was inserted into pLKO.1 vector. The target sequence of shRNA was shown as follows: shZFAS1: GGCTGAACCAGTTCCACAA, shMETTL3: GCTACAGATCCTGAGTTAG. To construct stable Hela cells with ZFAS1 overexpression, full-length ZFAS1 was subcloned into pLV plasmid. The above lentiviral expressing plasmids were co-transfected with the psPAX2 (Addgene) and pMD.2G plasmid (Addgene) into 293T cells using Turbofect (Thermo). After transfection, the lentiviral particles were harvested and used to infect CaSki or Hela cells. Forty-eight hours later, stable cells were selected using puromycin (3 μg/mL) for 1 week.
Transfection
The miR-647 mimics, inhibitor, and negative controls were purchased from RiboBio (Guangzhou, China). Cells were transfected using Lipofectamine 3000 reagent according to the manufacturer’s instructions. After 48 hours, cells were used for further experiments.
Isolation of Cytoplasmic and Nuclear RNA
The Cytoplasmic & Nuclear RNA Purification Kit (Norgen) was used to isolate and purify cytoplasmic and nuclear RNA as the manufacturer’s instructions. U6 was taken as an internal reference of nuclear RNA, while ACTB mRNA was taken as an internal reference of cytoplasmic RNA.
Real-Time PCR (RT-PCR)
Total RNA was extracted using the Trizol reagent according to the standard protocol. The cDNA was synthesized using PrimeScript RT reagent kit (TaKaRa). qRT-PCR analysis was carried out using the SYBR® Green Premix qPCR (Accurate Biotechnology, China) according to the manufacturer’s instructions. U6 and ACTB were used as internal controls. Primer sequences used are presented as follows:
ZFAS1-Forward: ACCAGTTCCACAAGGTTACTG,
ZFAS1-Reverse: CTTTATGCAGGTAGGCAGTTAGA,
METTL3-Forward: CACTGATGCTGTGTCCATCT,
METTL3-Reverse: CTTGTAGGAGACCTCGCTTTAC,
ACTB-Forward: GGACCTGACTGACTACCTCAT,
ACTB-Reverse: CGTAGCACAGCTTCTCCTTAAT.
Western Blot
Cells were collected and lysed with RIPA lysis buffer (Beyotime) on ice for 30 min. The concentration of protein samples was measured by using the Pierce BCA Protein Assay Kit (Thermo) according to the manufacturer’s instructions. Cell lysates were separated by SDS-polyacrylamide gel electrophoresis, and then transferred to polyvinylidene fluoride (PVDF) membranes (Millipore). The membranes were then blocked and incubated with the primary antibodies overnight at 4°C. The membranes were then incubated with corresponding secondary antibodies for 1 hour at room temperature. After wash for three times, the proteins were detected using Immobilon™ Western Chemiluminescent HRP Substrate (Millipore). Primary antibodies including anti-METTL3 (Abcam) and anti-β-actin (Proteintech) were used in this study.
Cell Counting Kit-8 (CCK-8) Assay
One thousand cells per well were suspended in 100 μL DMEM and seeded into 96-well plates. At different time point (0, 1, 2, 3, 4 days), 10 μL CCK-8 (Dojindo) solution was added into each well. After incubation for 2 hours, the optical density value was measured at 450 nm.
Colony Formation Assay
Five hundred cells per well were seeded in a 6-well plate and cultured for two weeks. Cell colonies were fixed with 4% paraformaldehyde for 30 min and stained with 0.5% crystal violet for 30 min at room temperature.
Transwell Assay
For migration and invasion detection, transwell chambers (Corning, USA) with a membrane pore size of 8 μm coated without or with Matrigel (BD Biosciences) were used, respectively. 1×105 or 2×105cells suspended in serum-free DMEM medium were seeded in the upper chambers, while DMEM medium supplemented with 10% FBS was added into the lower chambers. Twenty-four hours later, the cells were fixed by 4% paraformaldehyde, stained by 0.5% crystal violet, and finally counted under a microscope.
RNA Immunoprecipitation (RIP)
To validate the interaction between ZFAS1 and miR-647, cells were transfected with pCMV-MS2, pCMV-ZFAS1-MS2, pCMV-ZFAS1-mut-MS2 and pCMV-FLAG-MS2. Forty-eight hours later, cells were used to perform RIP assay using 5 μg anti-FLAG antibody (Sigma) and the Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (Millipore, Bedford, MA) according to the manufacturer’s instructions. To detect the interaction between METTL3 or AGO2 and ZFAS1, RIP assay was carried out by using 5 μg anti-METTL3 antibody (Abcam) or anti-AGO2 antibody (Abcam) and the Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit. IgG was taken as a negative control. The RNA fraction isolated and purified by RIP assay was then examined by RT-PCR.
RNA Pull-Down Assay
RNA pull-down assays were carried out using Pierce Magnetic RNA-Protein Pull-Down Kit (Thermo) according to the manufacturer’s instructions.
Luciferase Reporter Assay
The wild-type or mutant ZFAS1 fragment were subcloned into a luciferase reporter vector pmirGLO (Promega). The luciferase reporter plasmids were co-transfected into stable CC cells with miR-647 mimics or the negative control. The relative luciferase activity was measured with the Dual-Luciferase Reporter Assay System (Promega) as the manufacturer’s instructions. Data were presented as the relative ratio of firefly luciferase activity to renilla luciferase activity.
Methylated RNA Immunoprecipitation (MeRIP)
m6A modification of ZFAS1 was detected using Magna MeRIP™ m6A Kit (Millipore) as the manufacturer’s instructions. Briefly, 24 h after transfection, cells were harvested to perform RIP experiments using an m6A antibody. 1.5 μg anti-m6A antibody (Millipore) or anti-IgG (Cell Signaling Technology) was conjugated to protein A/G magnetic beads overnight at 4°C. A total of 100 μg RNA was then incubated with the antibody in IP buffer. Then, the purified RNA was analyzed via RT-PCR assay.
In vivo Model
To examine the effect of ZFAS1 on in vivo growth, nude mice were subjected to a subcutaneous injection of 5× 106 control and ZFAS1 silencing CaSki cells suspended in 0.2 mL DMEM medium. Tumor volume was monitored and calculated as (W2/2 ×L), where W and L represent the width and the length of the tumor, respectively. After 4 weeks, the mice were sacrificed, and the tumors were isolated and weighed. To examine the effect of ZFAS1 on in vivo metastasis, 1 × 106 control and ZFAS1 silencing CaSki cells were injected through the tail vein into the nude mice. After 8 weeks, the mice were sacrificed. The lung tissues were collected and the metastatic nodules were observed by hematoxylin and eosin (H&E) staining and counted under a microscope. All animal procedures were approved by the Medical Ethics Committee of Ningxia Medical University of Medicine and Institutional Animal Care. All animal experiments were performed according to approved protocols from the Institutional Animal Care and Use Committees and the national and institutional guidelines.
Statistical Analysis
Each experiment was repeated at least three times. All statistical analysis was carried out using SPSS 21.0 software (SPSS, Chicago, IL, USA). Student’s t-test was used for comparisons between two groups, and Variance analysis and a Tukey’s multiple comparisons test was applied for Comparisons among multiple independent groups (*P < 0.05, **P < 0.01, ***P < 0.001). The Kaplan–Meier curves and Log rank tests were used to evaluate the relationship between ZFAS1 expression and overall survival rate of CC patients. Correlation between ZFAS1 and miR-647 was analyzed with a Spearman rank correlation. P value less than 0.05 was considered to indicate statistical significance.
Results
Elevation of ZFAS1 Indicates Poor Clinical Outcome of Patients with CC
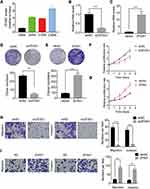
To determine ZFAS1 expression pattern in CC patients, the ZFAS1 expression between CC and matched adjacent nontumor tissues from 68 patients were analyzed using qRT-PCR assay. The results showed a significant upregulation of ZFAS1 expression in CC tissues compared to that in adjacent noncancerous tissues (Figure 1A). In order to investigate the clinical significance of ZFAS1 in CC, patients were sub-classified into high-ZFAS1 and low-ZFAS1 groups with the median of ZFAS1 expression in CC tissues as a cut-off value. As shown in Table 1, the high-ZFAS1 group had advanced FIGO stage, lymph node and distant metastasis. Next, we aim to determine the prognostic utility of ZFAS1 in CC. Survival analysis demonstrated that patients with high ZFAS1 expression had a significant worse overall survival compared to patients with low ZFAS1 expression (Figure 1B). These results suggested that upregulation of ZFAS1 could be an important event in the CC development and might act as an oncogene in CC.
 |
Table 1 The Correlation Analysis Between ZFAS1 Expression and Clinicopathological Features of CC Patients |
ZFAS1 Promotes CC Proliferation, Migration and Invasion in vitro
To explore the biological function of ZFAS1 in CC, we firstly detected the expression levels of different CC cell lines. Caski cells expressed the highest level of ZFAS1, while Hela cells had the lowest expression of ZFAS1 (Figure 2A). Therefore, ZFAS1 was stably depleted in Caski cells (Figure 2B) and overexpressed in Hela cells using lentiviral expressing system, respectively (Figure 2C). The results of CCK-8 and colony formation assays demonstrated that knockdown of ZFAS1 significantly attenuated the proliferation of Caski cells, whereas ectopic expression of ZFAS1 facilitated the proliferative ability of Hela cells (Figure 2DG).
The correlation between ZFAS1 and metastasis of CC indicated that ZFAS1 might affect the metastasis ability in CC. To validate the function of ZFAS1 in CC metastasis in vitro, transwell migration and invasion assays were utilized. It was observed that ZFAS1 silencing significantly suppressed whereas overexpression of ZFAS1 promoted the migration and invasion of CC cells (Figure 2H and I). These findings suggested that ZFAS1 facilitated CC cell proliferation, migration and invasion in vitro.
Knockdown of ZFAS1 Inhibits CC Growth and Metastasis of CC in vivo
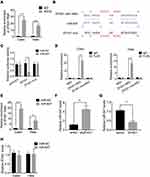
To further confirm the oncogenic function of ZFAS1 in vivo, control and ZFAS1-knockdown Caski cells were subcutaneously injected into nude mice. Both tumor volume and tumor weight in ZFAS1 knockdown group were much lower compared to that in control group (Figure 3A and B). Moreover, the results of tail vein injection models demonstrated that the number of pulmonary metastatic nodules formed by ZFAS1 knockdown group was significantly less than that in control group (Figure 3C). Altogether, these data confirmed that ZFAS1 knockdown restrained the tumorigenesis and metastasis of CC in vivo.
ZFAS1 Interacts with miR-647 in CC
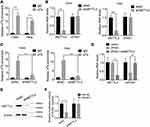
LncRNA usually acts as a competing endogenous RNA (ceRNA) to exert its regulatory functions. Since AGO2 is a core component of the RNA-induced silencing complex (RISC), we carried out a RIP assay using an anti-AGO2 antibody and observed a marked association of ZFAS1 with AGO2 (Figure 4A). This result suggested that ZFAS1 could act as a ceRNA. To screen the ZFAS1-targeted miRNAs, online prediction software LncBase (http://carolina.imis.athena-innovation.gr/diana_tools/web/) was used. Among the top 30 miRNAs, only miR-647 has been previously reported to show involvement in tumor progression.10–12 To validate this, we conducted luciferase reporter assays and found that transfection of miR-647 mimics reduced the luciferase activity of the wild-type ZFAS1 reporter gene, but not the mutant ZFAS1 (ZFAS1-mut) (Figure 4B and C). Moreover, we carried out a MS2-binding protein (MS2bp)-MS2-binding sequences (MS2bs)-based RIP (MS2-RIP) according to the previous studies.13,14 The results showed that ZFAS1 could interact with endogenous miR-647 in Hela and Caski cells (Figure 4D). Consistently, biotin-labeled miRNA pull-down assays showed markedly elevated ZFAS1 expression in Caski and Hela cells transfected with biotin-labeled miR-647 compared to that in the control (Figure 4E). Finally, it was observed that ZFAS1 negatively regulated the miR-647 expression (Figure 4F and G). We also detected the miR-647 expression in xenografts and found that knockdown of ZFAS1 markedly increased miR-647 expression (Supplemental Figure 1). Overexpression of miR-647 did not influence the ZFAS1 expression, suggesting that miR-647 associated with ZFAS1 but did not induce its degradation (Figure 4H). These results indicated that ZFAS1 acted as a ceRNA targeting miR-647.
ZFAS1 Sequestered miR-647 in a m6A-Dependent Manner
Recently, N(6)-methyladenosine (m6A) modification in mRNA or lncRNA functionally modulate the RNA splicing, localization, stability, and ceRNA activity.15,16 We performed methylated RNA immunoprecipitation (MeRIP) assays in Caski and Hela cells and found that ZFAS1 could be significantly enriched by m6A antibody compared to IgG (Figure 5A). The m6A modification sites of ZFAS1 was predicted by SRAMP online tool (www.cuilab.cn)(Supplemental Table 1). To further characterize the m6A methylation of ZFAS1, shRNA was used to knock down METTL3, a core component of m6A methylase complex. The downregulation of METTL3 could not change the expression levels of ZFAS1 (Figure 5B). Nevertheless, the m6A level of ZFAS1 was significantly decreased by depletion of METTL3, indicating that METTL3 is a major m6A methylase for ZFAS1 (Figure 5C).
Then, we investigated whether METTL3-mediated m6A modification of ZFAS1 influenced the ZFAS1-miR-647 interaction. We knocked down the METTL3 expression in ZFAS1-overexpressed Caski cells and found that METTL3 silence abolished the downregulation of miR-647 by ZFAS1 (Figure 5D). Moreover, we established a METTL3 knockdown Caski cells, and the knockdown efficacy was validated by Western blot assay (Figure 5E). Luciferase reporter assays in Control and METTL3 knockdown Caski cells transfected with the ZFAS1 reporter and miR-647 mimics. The results illustrated that the effect of miR-647 on the ZFAS1 reporter was abolished by METTL3 silence, as reporter activity showed no change after cotransfection with miR-647 miRNAs in METTL3 knockdown Caski cells (Figure 5F). These results revealed that the METTL3-mediated m6A modification of ZFAS1 was critical for the association between ZFAS1 and miR-647.
miR-647 is Partially Responsible for ZFAS1-Mediated Malignant Phenotypes of CC
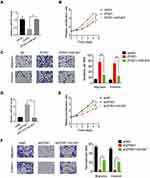
To investigate the involvement of miR-647 in ZFAS1-mediated malignant phenotypes of CC cells, rescue experiments were conducted. We transfected Hela cells stably overexpressing ZFAS1 with the miR-647 mimics (Figure 6A). Functional assays demonstrated that restoration of miR-647 expression partially rescued the promotive effects of ZFAS1 overexpression on cell proliferation, migration, and invasion (Figure 6B and C). Furthermore, Caski cells with ZFAS1 silence were transfected with miR-647 inhibitors (Figure 6D). Inhibition of miR-647 partially attenuated the suppressive effects of ZFAS1 knockdown in Caski cells (Figure 6E and F). These findings suggested that miR-647 contributes to CC cell proliferation, migration and invasion.
ZFAS1 Negatively Correlates with miR-647 Expression in CC Tissues
Finally, we examined the pathological correlation between ZFAS1 and miR-647. The expression of miR-647 in CC and adjacent normal tissues was evaluated using qRT-PCR. As shown in Figure 7A, miR-647 was significantly downregulated in CC tissues relative to adjacent noncancerous tissues. In addition, the negative correlation between ZFAS1 and miR-647 levels in CC tissues was observed, supporting that miR-647 was a target of ZFAS1 (Figure 7B).
Discussion
Increasing evidence suggests that lncRNAs play a critical role in the initiation and progression of human cancers.17 In this study, we found a significant upregulation of ZFAS1 expression in CC tissues, and the higher expression levels of ZFAS1 were associated with advanced FIGO stage, lymph node and distant metastasis, and poor clinical outcome in CC patients. Functionally, ZFAS1 promoted CC proliferation and metastasis in vitro and in vivo. Taken together, these results indicated that lncRNA ZFAS1 exerts oncogenic effects in CC.
ZFAS1 has been proved to act as a ceRNA in several kinds of human cancers. In HCC, ZFAS1 functions as an oncogene by binding miR-150 and abrogating its tumor-suppressive function.14 In colorectal cancer, lncRNA ZFAS1 sponges miR-484 to promote cell proliferation and invasion.18 ZFAS1 acts as a ceRNA to regulate miR-124, thereby elevating STAT3 expression and promoting esophageal squamous cell carcinoma progression.19 Additionally, ZFAS1 is highly expressed and promotes ovarian cancer cell proliferation, migration, invasion, and cisplatin resistance by sponging miR-548e and elevating CXCR4 expression.8 Here, we also identified miR-647 as a target of ZFAS1. ZFAS1 suppressed miR-647 expression and negatively correlated with miR-647 expression in CC tissues. Moreover, ZFAS1 exhibited oncogenic effects partially via miR-647, indicating that other mechanisms were involved in ZFAS1-mediated phenotypes of CC cells. miR-647 has been observed to abnormally express and act as a tumor-suppressive miRNA in some cancers. miR-647 targets HOXA9 to inhibit glioma cell proliferation and invasion.11 miR-647 promotes cisplatin-induced cell apoptosis via targeting IGF2 in lung cancer.20 Moreover, miR-647 targets SRF mRNA and represses the binding of SRF to MYH9 promoter, which attenuates the invasion and metastasis of gastric cancer.12 However, the downstream target mRNAs of miR-647 in CC remains unclear. The exact targets of ZFAS1-miR-647 need further exploration.
m6A is the most abundant internal RNA modification in mammalian cells and plays a crucial role in regulating RNA metabolism, such as RNA stability, localization, alternative splicing, translation and miRNA processing.21 Abnormal m6A modification of key oncogenes or tumor suppressors has been implicated in cancer occurrence and development. For example, m6A facilitates growth and progression of cervical and liver cancer via positively modulating the glycolysis of cancer cells. Mechanistically, the m6A modified 5ʹUTR of (pyruvate dehydrogenase kinase 4) PDK4 promotes its translation elongation and mRNA stability via binding with YTHDF1 and IGF2BP3.22 In oral squamous cell carcinoma (OSCC), METTL3 targeted the 3ʹ UTR of the oncogene c-Myc transcript to install the m6A modification, thereby enhancing its stability and OSCC tumorigenesis.23 m6A modification was also observed in lncRNAs transcript. The m6A modification mediated by METTL3 and METTL14 enhanced the stability of LNCAROD in head and neck squamous cell carcinoma cells.24 METTL3-mediated lncRNA FAM225A inhibits its degradation in nasopharyngeal cancer cells.25 A recent study demonstrated that m6A also modulate the interaction between lncRNA and miRNAs. Decreased m6A modification of linc1281 attenuated the interaction between linc1281 and let-7, suggesting that the m6A modification is necessary for the linc1281-mediated ceRNA model.15 Here, we found that METTL3 associated with ZFAS1, which increased the m6A level of ZFAS1. However, METTL3 did not affect the expression of ZFAS1. Knockdown of METTL3 abolished the miR-647-mediated suppression of luciferase activity of ZFAS1 reporter, indicating that ZFAS1 sequestered miR-647 in a m6A-dependent manner. To date, this is the first report to identify the m6A modification of ZFAS1 and characterize its role in CC.
Conclusion
In conclusion, we demonstrated ZFAS1 as an oncogenic lncRNA in CC. ZFAS1 promotes CC cell proliferation and metastasis by acting as a ceRNA that sponges miR-647, which is regulated by m6A modification. Our findings indicated that ZFAS1 may be a prognostic indicator and promising therapeutic target for CC patients.
Data Sharing Statement
The data and materials used in this study are available.
Ethics Approval and Consent to Participate
The informed consent was obtained from all patients in this research. The protocol of this study was approved by the Ethics Committee of General Hospital of Ningxia Medical University and performed in accordance with the World Medical Association Declaration of Helsinki.
Consent for Publication
Not applicable.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Funding
There is no funding to report.
Disclosure
The authors report no conflicts of interest for this work.
References
1. Givler DN, Givler A. Health Screening. Treasure Island (FL): StatPearls; 2020.
2. Wiltink LM, King M, Muller F, et al. A systematic review of the impact of contemporary treatment modalities for cervical cancer on women’s self-reported health-related quality of life. Supportive Care Cancer. 2020;28:4627–4644. doi:10.1007/s00520-020-05554-2
3. Gao Y, Wang JW, Ren JY, et al. Long noncoding RNAs in gastric cancer: from molecular dissection to clinical application. World J Gastroenterol. 2020;26(24):3401–3412.
4. Barth DA, Prinz F, Teppan J, Jonas K, Klec C, Pichler M. Long-Noncoding RNA (lncRNA) in the regulation of hypoxia-inducible factor (HIF) in Cancer. Non-Coding RNA. 2020;6:3. doi:10.3390/ncrna6030027
5. Amelio I, Bernassola F, Candi E. Emerging roles of long non-coding RNAs in breast cancer biology and management. Semin Cancer Biol. 2020. doi:10.1016/j.semcancer.2020.06.019
6. Singh D, Khan MA, Siddique HR. Emerging role of long non-coding RNAs in cancer chemoresistance: unravelling the multifaceted role and prospective therapeutic targeting. Mol Biol Rep. 2020;47:5569–5585. doi:10.1007/s11033-020-05609-x
7. Duan R, Li C, Wang F, Han F, Zhu L. The long noncoding RNA ZFAS1 potentiates the development of hepatocellular carcinoma via the microRNA-624/MDK/ERK/JNK/P38 signaling pathway. Onco Targets Ther. 2020;13:4431–4444. doi:10.2147/OTT.S246278
8. Zhang J, Quan LN, Meng Q, et al. miR-548e sponged by ZFAS1 regulates metastasis and cisplatin resistance of OC by targeting CXCR4 and let-7a/BCL-XL/S signaling axis. Mol Ther Nucleic Acids. 2020;20:621–638. doi:10.1016/j.omtn.2020.03.013
9. Askarian-Amiri ME, Crawford J, French JD, et al. SNORD-host RNA Zfas1 is a regulator of mammary development and a potential marker for breast cancer. Rna. 2011;17(5):878–891.
10. Qian C, Liao CH, Tan BF, et al. LncRNA PROX1-AS1 promotes proliferation, invasion, and migration in prostate cancer via targeting miR-647. Eur Rev Med Pharmacol Sci. 2020;24(6):2938–2944.
11. Qin K, Tian G, Chen G, Zhou D, Tang K. miR-647 inhibits glioma cell proliferation, colony formation and invasion by regulating HOXA9. J Gene Med. 2020;22(3):e3153. doi:10.1002/jgm.3153
12. Ye G, Huang K, Yu J, et al. MicroRNA-647 Targets SRF-MYH9 axis to suppress invasion and metastasis of gastric cancer. Theranostics. 2017;7(13):3338–3353. doi:10.7150/thno.20512
13. Yuan JH, Yang F, Wang F, et al. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25(5):666–681. doi:10.1016/j.ccr.2014.03.010
14. Li T, Xie J, Shen C, et al. Amplification of long noncoding RNA ZFAS1 promotes metastasis in hepatocellular carcinoma. Cancer Res. 2015;75(15):3181–3191. doi:10.1158/0008-5472.CAN-14-3721
15. Yang D, Qiao J, Wang G, et al. N6-Methyladenosine modification of lincRNA 1281 is critically required for mESC differentiation potential. Nucleic Acids Res. 2018;46(8):3906–3920. doi:10.1093/nar/gky130
16. Shen H, Lan Y, Zhao Y, Shi Y, Jin J, Xie W. The emerging roles of N6-methyladenosine RNA methylation in human cancers. Biomarker Research. 2020;8:24. doi:10.1186/s40364-020-00203-6
17. Luo F, Wen Y, Zhou H, Li Z. Roles of long non-coding RNAs in cervical cancer. Life Sci. 2020;117981.
18. Xie S, Ge Q, Wang X, Sun X, Kang Y. Long non-coding RNA ZFAS1 sponges miR-484 to promote cell proliferation and invasion in colorectal cancer. Cell Cycle. 2018;17(2):154–161. doi:10.1080/15384101.2017.1407895
19. Li Z, Qin X, Bian W, et al. Exosomal lncRNA ZFAS1 regulates esophageal squamous cell carcinoma cell proliferation, invasion, migration and apoptosis via microRNA-124/STAT3 axis. J Experimental Clinical Cancer Res. 2019;38(1):477. doi:10.1186/s13046-019-1473-8
20. Jiang W, Zhao X, Yang W. MiR-647 promotes cisplatin-induced cell apoptosis via downregulating IGF2 in non-small cell lung cancer. Minerva Med. 2019.
21. Zhu ZM, Huo FC, Pei DS. Function and evolution of RNA N6-methyladenosine modification. Int J Biol Sci. 2020;16(11):1929–1940. doi:10.7150/ijbs.45231
22. Li Z, Peng Y, Li J, et al. N(6)-methyladenosine regulates glycolysis of cancer cells through PDK4. Nat Commun. 2020;11(1):2578. doi:10.1038/s41467-020-16306-5
23. Zhao W, Cui Y, Liu L, et al. METTL3 facilitates oral squamous cell carcinoma tumorigenesis by enhancing c-myc stability via YTHDF1-mediated m(6)A modification. Mol Ther Nucleic Acids. 2020;20:1–12. doi:10.1016/j.omtn.2020.01.033
24. Ban Y, Tan P, Cai J, et al. LNCAROD is stabilized by m6A methylation and promotes cancer progression via forming a ternary complex with HSPA1A and YBX1 in head and neck squamous cell carcinoma. Mol Oncol. 2020;14(6):1282–1296. doi:10.1002/1878-0261.12676
25. Zheng ZQ, Li ZX, Zhou GQ, et al. Long noncoding RNA FAM225A promotes nasopharyngeal carcinoma tumorigenesis and metastasis by acting as ceRNA to sponge miR-590-3p/miR-1275 and upregulate ITGB3. Cancer Res. 2019;79(18):4612–4626. doi:10.1158/0008-5472.CAN-19-0799
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.