Back to Journals » The Application of Clinical Genetics » Volume 14
Y-Chromosome Microdeletions: A Review of Prevalence, Screening, and Clinical Considerations
Authors Rabinowitz MJ, Huffman PJ, Haney NM, Kohn TP
Received 16 November 2020
Accepted for publication 18 January 2021
Published 12 February 2021 Volume 2021:14 Pages 51—59
DOI https://doi.org/10.2147/TACG.S267421
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Martin Maurer
Matthew J Rabinowitz, Phillip J Huffman, Nora M Haney, Taylor P Kohn
The James Buchanan Brady Urological Institute, Johns Hopkins University School of Medicine, Baltimore, MD, USA
Correspondence: Taylor P Kohn
James Buchanan Brady Urological Institute, Johns Hopkins University School of Medicine, 600 N. Wolfe Street/Marburg 134, Baltimore, MD, 21287, USA
Email [email protected]
Abstract: Deletions within the male-specific region of the Y-chromosome, known as Y-Chromosome Microdeletions (YCMs), are present in as many as 5% and 10% of severe oligospermic and azoospermic men, respectively. These microdeletions are distinguished by which segment of the Y chromosome is absent, identified as AZFa (the most proximal segment), AZFb (middle), and AZFc (distal). The reported prevalence of YCMs within the world’s populations of infertile men displays vast heterogeneity, ranging from less than 2% to over 24% based on region and ethnicity. AZFc is the most commonly identified YCM, and its phenotypic presentation provides for the highest chance for fertility through artificial reproductive techniques. Conversely, deletions identified in the subregions of AZFa, AZFb, or any combination of regions containing these segments, are associated with low probabilities of achieving pregnancy. A putative mechanism explaining this discrepancy lies within the expression of autosomal, DAZ-like genes which could serve to “rescue” wild type AZFc gene expression and hence spermatogenesis. Nevertheless, recent reports challenge this dogma and stress the importance of further analysis when an AZFb deletion is detected. The screening thresholds to determine which oligospermic and azoospermic men are tested for potential YCMs has been recently contested. More recent literature supports lowering the threshold from 5 million sperm/mL of ejaculate to 1 million/mL as the frequency of YCMs in men with sperm concentrations between 1 and 5 million sperm/mL is very low (∼ 0.8%). As such, subsequent guidelines should recommend a lower screening threshold. While YCMs are extremely common globally, the understanding of their clinical significance in the field remains scattered and without consensus. Furthermore, very little is currently known about partial deletions within the AZFc region, such as b1/b3, b2/b3, and gr/gr. Hence, this review aimed to summarize and discuss modern trends in the epidemiology, screening guidelines, and clinical considerations pertaining to YCMs.
Keywords: Y chromosome, abnormalities, chromosomal, gene deletion, genetics, infertility, male
Introduction
Infertility is defined as the inability to conceive after 12 months of unprotected intercourse or 6 months of unprotected sex in the setting of advanced maternal age.1 It is estimated that infertility will affect one of every 12 men in their lifetimes. Of men presenting with infertility, 90% will have an underlying oligospermia or azoospermia.2 Oligospermia is defined as a sperm concentration of less than 15 million sperms per milliliter of ejaculate, below the 5th percentile of fertile men;3 this classification can be further stratified into mild, moderate, and severe categories defined as 10–15 million sperm/mL, 5–10 million sperms/mL, and any sperm to 5 million sperm/mL, respectively.4 Azoospermia is the complete absence of sperm in the ejaculate and is stratified into two classifications: obstructive and non-obstructive. Obstructive azoospermia is classified as a post-testicular obstruction of the ductal system impeding the movement of sperm at any location in the reproductive tract – sperm is classically seen on testicular biopsy. Conversely, non-obstructive azoospermia (NOA) is defined as the absence of sperm in the ejaculate due to primary testicular failure and is commonly diagnosed by testicular biopsy suggestive of spermatogenic failure.5
One of every four cases of non-obstructive azoospermia is presumed to be caused by an underlying genetic defect. Common, non-genetic causes of NOA include cryptorchidism, varicocele, or testicular neoplasia; nonetheless, the underlying cause of many cases of non-obstructive azoospermia remains idiopathic.6 The genetic bases of primary testicular failure have been the subject of much investigation in the field of male infertility, particularly deletions identified on autosomal and Y sex chromosomes.7 The Y chromosome is classically divided into two sections; the pseudoautosomal regions and the male-specific region. Pseudoautosomal regions (PAR1 and PAR2) are classified as areas of the chromosome that may recombine with the X chromosome during meiosis and account for about 5% of the Y chromosome.8
The male-specific region of the Y Chromosome, abbreviated as MSY, accounts for the additional 95% of the chromosome that does not recombine or crossover with the X chromosome during meiosis. The MSY region has been conserved in primate and eutherian genomes for millions of years, made up in part by amplicon sequences which are expressed almost exclusively in the testis. About 25% of the MSY euchromatin in man is constituted by such amplicon regions organized into eight massive palindromic sequences that are virtually identical. These amplicon sequences undergo frequent gene conversion (non-reciprocal transfer) which has served as a mechanism to prevent their genetic decay. These large amplicon regions are suspectable to homologous recombination that could therefore result in deletions within the MSY region.8,9
It is assumed that the genes contained in the MSY are integral to spermatogenesis through myriad functions including transcription, gene silencing, ubiquitination, and maintenance of microtubule networks. It should not be surprising, then, that the most frequent known genetic causes of impaired spermatogenesis, accounting for as much as 5% and 10% of severe oligospermic and azoospermic men, respectively, are stereotypical deletions within the MSY region known as Y-chromosome microdeletions.10 Moreover, such deletions, enabled by the homologous recombination of highly conserved amplicon regions, have been studied extensively throughout many placental mammalian genomes including horse, chimpanzee, bonobo, and various bovine species. These genomic studies have widely yielded similar findings to those in man and emphasize the vital role MSY plays in spermatogenesis.11–14
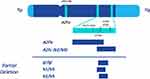
In 1976 a conserved, terminal deletion within MSY located at the long arm (11q) was identified in six azoospermic men. Based on the terminal deletions in this series and their corresponding phenotypes, it was hypothesized that the 11q region of the Y-chromosome performed vital functions in spermatogenesis and was termed the azoospermia factor (AZF).15 More than 10 years after the identification of AZF, “microdeletions” at 76 discrete DNA loci were mapped into three subregions of Yq11 based on their presentation in azoospermic men as well as their putative action in different phases of male germ cell development; they were termed AZFa (the most proximal segment), AZFb (middle), and AZFc (most distal) (Figure 1).16
These deletions within AZF have therefore been categorized as Y-chromosome microdeletions (YCMs). The putative mechanism behind the synthesis of YCMs, as stated previously, involves amplicon sequences located within the MSY making certain segments of the region suspectable to deletions and duplications via non-allelic homologous recombination.17 Moreover, several partial deletions in regions of AZFc, such as gr/gr and b2/b3, have been frequently identified in men and will also be discussed herein.18
While Y-chromosome microdeletions and the genetic basis of male-factor infertility have emerged as critical areas of study in male fertility, the understanding of their clinical significance in the field remains scattered and without consensus. Our review, therefore, aims to summarize and discuss the current body of literature pertaining to YCMs, focusing on trends in epidemiology, screening guidelines, and clinical considerations.
Prevalence and Presentation of Y-Chromosome Microdeletions
For more than two decades, YCMs have been reported across the globe using reliable duplex and/or multiplex PCR diagnostic testing. Despite this breadth of research, reporting pertaining to the prevalence and presence of YCMs remains highly heterogenous. According to the Practice Committee of the American Society for Reproductive Medicine (ASRM), the prevalence of YCMs might be as high as 2% in the general population of unselected men.19 The European Academy of Andrology (EAA) defines the prevalence of YCMs in unselected men to be far lower, around 1 in 4000 (0.025%).17
Genetic screening studies have informed the understanding that YCMs are extremely rare in men who are normospermic, defined as having a sperm concentration greater than 20 million per mL of ejaculate. A 2001 study found no Yq deletions amongst 392 normospermic men screened.20 Further, a recent meta-analysis of over 10,000 North American men found that while 5% of severely oligospermic patients (defined in this study as containing less than 1 million sperms per mL of ejaculate) had an AZF microdeletion, the prevalence of these same deletions was less than 1% amongst men with greater than 20 million sperm/mL of ejaculate (normospermic concentrations).21 Hence, it is believed full deletions in the AZF regions are specific for spermatogenic failure. These findings emphasize the distinction between fertility and normospermia, which are not synonymous. While oligospermic men may be fertile and never be diagnosed with a low sperm concertation or a YCM, normospermia is exclusively defined by semen parameters (and therefore the quality of spermatogenesis) rather than a man’s fertility.
In accordance with the proposed role AZF plays in spermatogenesis, YCMs are far more prevalent amongst infertile men and/or men with abnormal semen parameters. The Practice Committee of the ASRM estimated in 2015 that the prevalence of YCMs amongst men with severe oligospermia or azoospermia is as high as 16%. A 2016 review by the European Association of Urology reported the prevalence of YCMs to be 8–12% in azoospermic men and approximately 3–7% in men with severe oligospermia.22 It is important to highlight that the small prevalence of YCMs in the unselected population is likely constituted by men who are in fact oligospermic or azoospermic rather than silent mutations amongst men who are normospermic, which is extremely unlikely as described above.
The reported presence of YCMs within populations of infertile men displays vast heterogeneity that has been outlined by geographical region and ethnicity. Estimates of the worldwide frequencies of AZF deletions in infertile men vary greatly, ranging from 12% in the United States and 24.2% in Iran to less than 2% in countries such as Germany and Austria. Even to the casual observer, there appears no obvious geographical pattern accounting for which parts of the world correspond to which frequencies.23 Moreover, a 2019 study screened 1030 infertile Japanese men for YCMs, the first large-scale study to assess such genetics in the region. This analysis yielded a prevalence of nearly 7% (including all AZF rearrangements) in their population made up almost exclusively of severely oligospermic or azoospermic men. Amongst the various AZF deletions screened for in this study, AZFc was the region most commonly lost.24
Screening infertile and subfertile men to determine the presence of an underlying Y-chromosome microdeletion has proven to be vital in the clinical setting, as the extent of such deletions often informs clinical decision-making and recommendations for artificial reproductive technologies (ART). Between AZFa, AZFb, and AZFc deletions, the latter of the three is the most frequently diagnosed YCM, accounting for 60–80% of all reported deletions.17,25 Fortunately, while AZFc is the most salient microdeletion, it also the microdeletion associated with the highest probability of success through ART.
For example, microdissection testicular sperm extraction (mTESE) is considered the “gold standard” of treatment for men with non-obstructive azoospermia. The mTESE is a procedure that involves bivalving of the testicle and searching for and collecting dilated seminiferous tubules (believed to be predictive of ongoing spermatogenesis) using an operative microscope. For use in men with non-obstructive azoospermia, the mTESE has a successful sperm retrieval rate of approximately 52% over all populations.26 However, for men with YCMs, the rate of successful sperm retrieval, and therefore the preoperative probability of success, depends greatly on the region of AZF that is absent. Deletions in the AZFc region of the MSY are associated with the highest retrieval rates of any microdeletion discussed herein, reported between 50% and 80% in past studies.17,25,27 Unfortunately, complete YCMs detected in the AZFa and AZFb regions have been linked with extremely unfavorable sperm retrieval rates and clinical outcomes. Further, deletions containing sections of multiple AZF regions, such as AZFbc and AZFabc, have also been associated with the same poor sperm retrieval.17,24,25,28
Nonetheless, recent reports have identified very few men, originally diagnosed with full AZFb deletions using basic genetic testing, to contain residual sperm production, challenging the formally discussed understanding of such YCMs. These reports, while limited, suggest that some men who are originally diagnosed with AZFb deletions might have a chance for successful sperm retrieval. The authors of one such report identify an extended genetic analysis, using a specialized sY1192 primer as a decision-marker, to determine the true extent of the AZFb deletion and therefore better elucidate the man’s potential for successful sperm retrieval; if sY1192 is present, the authors’ contest ART might still be a viable option for the patient.29
A proposed mechanism for the disparate outcomes associated with deletions of AZFa, AZFb, AZFc, AZFbc, and AZFabc focuses on the importance of the DAZ gene, which is a copy number variant (CNV) located in four nearly identical copies in the AZFc reference sequence (DAZ1, DAZ2, DAZ3, and DAZ4).30,31 While it is understood that most genes contained within AZF are critical to successful spermatogenesis, it is believed that two additional, analogous copies of the DAZ gene, known as DAZL (Daz-Like), are located on the autosomal chromosome 3, in addition to its copies contained in AZFc. Unlike the other YCMs discussed, when AZFc alone is absent, it is proposed that the autosomal gene copies of DAZL can serve to “rescue” spermatogenesis and therefore facilitate the production of rare sperm required for retrieval via mTESE.25,27,32–35
Hence, distinguishing not only the presence of a potential Y-chromosome microdeletion but also the extent of its deletion, is paramount in clinical decision-making involving ART and infertility counseling. Professional societies including the European Academy of Andrology and the American Society for Reproductive Medicine both recommend testing men with oligospermia or non-obstructive azoospermia for YCMs.17,19 Based on the poor outcomes previously discussed, sperm retrieval procedures are not recommended for patients with a verified presence of a deletion in the regions of AZFa, AZFb, AZFabc, or AZFbc, although the methods through which to verify AZFb deletions have been brought to question by limited reports previously discussed.17,29
Even while men containing AZFc deletions may be recommended for ART following the genetic screening, there are further clinical considerations. Any male offspring of a man with an AZFc deletion will inherit their father’s Y chromosome with microdeletions. Hence, barring unforeseen future breakthroughs allowing for genetic restorations, these male children will be equally or more severely infertile than their fathers.36 Additionally, screening of male relatives and karyotyping is recommended as AZFc microdeletions have been implicated in higher risks of Turner syndrome (45, X0) and ambiguous genitalia in any viable male offspring. For this reason, genetic counseling of men with a verified presence of an AZFc deletion should be considered mandatory.36 For men with an AZFc deletion who wishes to proceed with ART, couples may opt for use of antenatal testing, such as preimplantation genetic testing for aneuploidy (PGT-A), in order to select for female progeny for embryo transfer (thus not passing on this deletion), or use donor sperm.21
Screening for YCM in Oligospermic Men
Screening for YCM in oligospermic men is performed through PCR amplification. According to the European Academy of Andrology (EAA) and European Molecular Genetics Quality Network (EMQN) guidelines, the PCR reaction should utilize sequence-tagged sites (STS) primers surrounding regions of the Y chromosome that are non-polymorphic and commonly deleted in oligospermic and azoospermic men. To promote sensitivity and specificity of the analysis, two STS loci should be amplified in each AZF region. The following primers are recommended by the EAA/EMQN guidelines (Figure 1):17
- For AZFa: sY84, sY86
- For AZFb: sY127, sY134
- For AZFc: sY254, sY255 (both in the DAZ gene)
Superfluous STS primers should not be used as they do not have clinical significance and can in fact cause confusion in the interpretation of the results.37 Further, though whole genome sequencing (WGS) has risen in popularity, it is currently not recommended for YCM screening. Most deleterious YCMs are widely dispersed along the amplicon regions of the MSY euchromatin. WGS relies on short reads, which has shown to result in the filtering of the mutation sites upon sequencing. The use of PCR amplification to screen these STS sites is more appropriate to identify these deletions.38,39
Detection of a deletion in one or multiple of these loci may require further analysis using more specific primers to determine the extension (partial or complete) of the deletion. For example, previously recorded partial AZFa deletions have demonstrated milder phenotypes than full deletions, while full deletions of AZFa are incompatible with ART as discussed previously.40 AZFb and AZFbc deletions should be identified as partial or complete, as there is higher availability for sperm cells in ART in men with partial deletions.41 Further, limited recent reports endorse the use of additional STS primers in order to confirm previously diagnosed complete AZFb deletions (ie sY1192 as discussed previously).29 Complete deletions of AZFbc should also be studied using specific markers to differentiate between P5/distal P1 and P4/distal P1 deletions.42 In the case of a complete deletion of AZFc or AZFbc, screening for heterochromatin marker sY160 is indicated to differentiate b2/b4 deletions from terminal deletions. The latter are characterized by an absence of sY160, which are associated with a mosaic karyotype (46,XY/45,X). The 45,X karyotype has been previously shown to be a negative prognostic factor for normal spermatogenesis and should therefore be considered in screening protocols.43
Ideally, the reactions should be performed in multiplex format to include relevant positive (SRY) and negative (ZFX/ZFY X-chromosome genes) internal controls. A female sample and a blank (replacing DNA with water) sample should also be analyzed in parallel as controls for DNA contamination. This set of primers and quality control methods have demonstrated reproducible and accurate results, but repetitive testing may be done in the scenario of inconclusive results or suspected technical failure. Incorporation of these guidelines, laboratory standardization and continued evaluation via annual external quality assessment (EQA/EMQN) schemes have reduced the diagnostic error from approximately 8% to 1–2% (as of 2014).17
Several reports also have identified YCMs in germline, sperm DNA, which are hypothesized to be more prevalent than in genomic DNA, derived from lymphocytes, due to the increased oxidation stress germline DNA is subjected to. Such deletions, if confirmed to be occurred at higher rates in sperm DNA, could dictate the source of DNA most useful to assay for men who wish to pursue ART. Unfortunately, these reports do not establish the extent or origin of the YCMs identified in sperm DNA and lack statistical power due to their small sample sizes. The collection of blood-derived DNA hence remains the most cost-effective and widely available method for YCM screening.44–47
The issue of which males should be screened for YCMs remains a controversial topic. Published guidelines by professional societies such as the ASRM and the EAA/EMQN recommend screening for men with severe oligospermia (<5-0million sperm/mL ejaculate).17,19 However, several recent studies have suggested that this cutoff may be arbitrary and cost-ineffective for multiple reasons. First, the current cutoff for severe oligospermia may not reflect a clinically relevant threshold for screening. Meta-analysis by Kohn et al found no significant difference in complete YCM prevalence between males with sperm counts >1–5 million/mL and >5–20 million/mL (0.8% [95% CI: 0.5–1.3%] vs 0.5% [95% CI: 0.2–0.9%], P=0.14). There was, however, a significant difference in complete YCM prevalence between males with sperm counts >0–1 million/mL and >1–5 million/mL (5.0% [95% CI: 3.6–6.8%] vs 0.8% [95% CI: 0.5–1.3%], P<0.001). Thus, they suggest lowering the threshold for genetic testing to 1 million sperm/mL.21
A second reason to continuously evaluate, and potentially lower, the YCM screening threshold is the decrease in average sperm count over time. A 2017 meta-analysis of 185 studies and 42,935 semen samples showed a significant decrease in average sperm concentration in men from North America, Europe, and Australia during 1973–2011.48 This finding was corroborated by a second 2017 meta-analysis that found a 57% decline in average sperm concentration in men from North America, Europe, Asia, and Africa during 1980–2015.49 Fluctuations in average sperm concentration indicate the need for further evaluation of the screening threshold to reflect the current landscape of men’s reproductive health and minimize superfluous screening.
Lowering the threshold could greatly reduce costs without sacrificing sensitivity. A retrospective study of 1473 men seeking fertility evaluation by Johnson et al found that a 5 million sperm/mL threshold for screening has a 100% sensitivity and a 13% specificity. A theoretical reduction in this threshold to 1 million sperm/mL still had a 100% sensitivity, but specificity increased to 24%.50 A further reduction to 0.5 million sperm/mL increases specificity again, but this threshold is not generally recommended due to the relatively high prevalence of YCMs in men with sperm counts >0.5–1million sperm/mL.50,51 YCM testing costs approximately $250 out-of-pocket per patient, so limiting excessive testing could reduce medical costs.52 Kohn et al estimated that this reduced testing threshold could lower the rate of testing by 42%, saving approximately $1,107,054 annually.21 Another recent study of 3023 oligospermic men (<5 million sperm/mL) found a reduced screening threshold of 1 million sperm/mL could increase testing specificity by 22.1% without a significant decrease in sensitivity. This reduced the testing rate in the study site by 23.1%, saved the hospital $48,225 over 5 years, and minimized subjecting patients to the psychological stress of testing.53 Ultimately, the question of who should be screened for YCMs is an important one, and current studies suggest that more investigation and debate are needed to find a cost-effective and medically appropriate threshold.
Partial AZFc Deletions and Duplications
The primary presentation of YCMs is classified as a “complete” deletion of an entire AZF region, with AZFc being the most commonly deleted region. The AZFc region is also susceptible to partial deletions and duplications, in which one of the three specific sections of AZFc (b1/b3, b2/b3, and gr/gr) are deleted or duplicated through non-allelic homologous recombination (NAHR). Both partial duplications and deletions have shown to be risk factors for impaired spermatogenesis and male infertility, though there are much fewer studies on duplications.54,55 The most clinically important partial AZFc genetic variation is gr/gr deletion, in which a deletion of the proximal half of the AZFc region can result in a reduction in sperm count.56
The spermatogenic phenotypes of gr/gr carriers have been shown to be highly variable between studies, ethnicities, and geographic regions. There are consistent data indicating an association between gr/gr deletions and impaired spermatogenesis in certain populations, such as Italian and Spanish Caucasians.57–59 Another 2012 study of over 20,000 men from India, Poland, Tunisia, Vietnam, and the United States showed that gr/gr deletions doubled the risk of severe oligospermia.60 Contrarily, a 2020 study of 30 Bulgarian men with severe oligospermia (>0–5 million sperm/mL) showed no association between this deletion and male infertility.61 There was also no association between gr/gr deletion and oligo-/azoospermia found in a 2005 study of 348 German men with non-obstructive oligo-/azoospermia.62 Additionally, some studies have pointed to gr/gr duplication as a significant risk factor for male infertility in Asian men (OR = 2.71, 95% CI = 1.38‐5.32, P= 0.004), but not European men.63 This variability in results has precluded a consensus on the spermatogenic effects of partial YCMs. Furthermore, there has been controversy surrounding the methodologies (eg selection biases, screening techniques) of studies focusing on partial deletions. This stark lack of decisive work has made reaching a screening consensus difficult.17
Despite this, there may be some clinical benefits to screening for partial YCMs. For example, there has been evidence that partial AZFc deletions may expand and increase the risk of complete AZFc deletion in subsequent generations, which would undoubtedly result in spermatogenic impairment of the offspring.64 Partial AZFc (gr/gr) deletions may also predispose men to testicular germ cell tumors, particularly in normospermic carriers.65 Screening for partial AZFc deletions, such as gr/gr, may help prevent these negative clinical outcomes at little additional cost. This practice may be particularly beneficial in certain populations (ie Spanish and Italian men), in which previous research has consistently shown associations between gr/gr and infertility. Still, current guidelines do not recommend screening for partial YCMs given the ambiguity associated with the associated phenotype.17
Conclusion
Y-Chromosome Microdeletions identified within the AZF gene are highly suggestive of spermatogenic failure, irregular sperm concentration, and male factor infertility. Despite a growing awareness of YCMs within the world’s infertile male population, the true prevalence and implications of such rearrangements remain highly heterogenous. The clinical presentations of YCMs differ by which subregion of the AZF is deleted; AZFc deletions have been demonstrated to have the best clinical outcomes for fertility, while deletions of AZFa, AZFb, AZFbc, and AZFabc all have extremely poor outcomes and should not undergo sperm retrieval procedures. Recent literature endorses lowering the threshold for screening from 5 million sperm/mL to 1 million/mL as the frequency of YCMs in men with sperm concentrations between 1 and 5 million sperm/mL is very low (0.8%). Other factors, including ethnicity and temporal trends in sperm concentration, may also contribute to the likelihood of finding a YCM. As such, subsequent guidelines should recommend updated screening criteria that include a lower screening threshold and consideration of other variables to more appropriately designate patients for screening.
Disclosure
The authors report no conflicts of interest for this work.
References
1. World Health Organization, editor. WHO Laboratory Manual for the Examination and Processing of Human Semen.
2. Kumar N, Singh A. Trends of male factor infertility, an important cause of infertility: a review of literature. J Hum Reprod Sci. 2015;8(4):191. doi:10.4103/0974-1208.170370
3. Cooper TG, Noonan E, von Eckardstein S, et al. World Health Organization reference values for human semen characteristics*‡. Hum Reprod Update. 2010;16(3):231–245. doi:10.1093/humupd/dmp048
4. Hopps CV, Schlegel PN, Goldstein M. 49 - A practical approach to male infertility. In: Legato MJ, editor. Principles of Gender-Specific Medicine. Academic Press; 2004:538–549. doi:10.1016/B978-012440905-7/50318-2.
5. Cocuzza M, Alvarenga C, Pagani R. The epidemiology and etiology of azoospermia. Clinics (Sao Paulo). 2013;68(Suppl 1):15–26. doi:10.6061/clinics/2013(Sup01)03
6. Chiba K, Enatsu N, Fujisawa M. Management of non‐obstructive azoospermia. Reprod Med Biol. 2016;15(3):165–173. doi:10.1007/s12522-016-0234-z
7. Kim SY, Lee BY, Oh AR, Park SY, Lee HS, Seo JT. Clinical, hormonal, and genetic evaluation of idiopathic nonobstructive azoospermia and klinefelter syndrome patients. CGR. 2017;153(4):190–197. doi:10.1159/000487039
8. Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423(6942):825–837. doi:10.1038/nature01722
9. Li G, Davis BW, Raudsepp T, et al. Comparative analysis of mammalian Y chromosomes illuminates ancestral structure and lineage-specific evolution. Genome Res. 2013;23(9):1486–1495. doi:10.1101/gr.154286.112
10. Krausz C, Casamonti E. Spermatogenic failure and the Y chromosome. Hum Genet. 2017;136(5):637–655. doi:10.1007/s00439-017-1793-8
11. Janečka JE, Davis BW, Ghosh S, et al. Horse Y chromosome assembly displays unique evolutionary features and putative stallion fertility genes. Nat Commun. 2018;9(1):2945. doi:10.1038/s41467-018-05290-6
12. Oetjens MT, Shen F, Emery SB, Zou Z, Kidd JM. Y-chromosome structural diversity in the bonobo and chimpanzee lineages. Genome Biol Evol. 2016;8(7):2231–2240. doi:10.1093/gbe/evw150
13. Lahn BT, Pearson NM, Jegalian K. The human Y chromosome, in the light of evolution. Nat Rev Genet. 2001;2(3):207–216. doi:10.1038/35056058
14. Das PP, Krishnan G, Doley J, et al. Identification and expression profiling of MSY genes of yak for bull fertility. J Genet. 2019;98(2). doi:10.1007/s12041-019-1091-4.
15. Tiepolo L, Zuffardi O. Localization of factors controlling spermatogenesis in the nonfluorescent portion of the human y chromosome long arm. Hum Genet. 1976;34(2):119–124. doi:10.1007/BF00278879
16. Vog PH, Edelmann A, Kirsch S, et al. Human Y chromosome azoospermia factors (AZF) mapped to different subregions in Yq11. Hum Mol Genet. 1996;5(7):933–943. doi:10.1093/hmg/5.7.933
17. Krausz C, Hoefsloot L, Simoni M, Tüttelmann F; European Academy of Andrology, European Molecular Genetics Quality Network. EAA/EMQN best practice guidelines for molecular diagnosis of Y-chromosomal microdeletions: state-of-the-art 2013. Andrology. 2014;2(1):5–19. doi:10.1111/j.2047-2927.2013.00173.x
18. Navarro-Costa P, Pereira L, Alves C, et al. Characterizing partial AZFc deletions of the Y chromosome with amplicon-specific sequence markers. BMC Genomics. 2007;8:342. doi:10.1186/1471-2164-8-342
19. Practice Committee of the American Society for Reproductive Medicine. Diagnostic evaluation of the infertile male: a committee opinion. Fertil Steril. 2015;103(3):e18–e25. doi:10.1016/j.fertnstert.2014.12.103
20. Krausz C, Forti G, McElreavey K. The Y chromosome and male fertility and infertility1. Int J Androl. 2003;26(2):70–75. doi:10.1046/j.1365-2605.2003.00402.x
21. Kohn TP, Kohn JR, Owen RC, Coward RM. The prevalence of Y-chromosome microdeletions in oligozoospermic men: a systematic review and meta-analysis of European and North American studies. Eur Urol. 2019;76(5):626–636. doi:10.1016/j.eururo.2019.07.033
22. Jungwirth A, Diemer T, Feng Z, Krausz C, Minhas S, Tournaye H. EAU Guidelines on Male Infertility. EAU Guidelines Office; 2018.
23. Simoni M, Tüttelmann F, Gromoll J, Nieschlag E. Clinical consequences of microdeletions of the Y chromosome: the extended Münster experience. Reprod Biomed Online. 2008;16(2):289–303. doi:10.1016/S1472-6483(10)60588-3
24. Iijima M, Shigehara K, Igarashi H, et al. Y chromosome microdeletion screening using a new molecular diagnostic method in 1030 Japanese males with infertility. Asian J Androl. 2019;22(4):368–371. doi:10.4103/aja.aja_97_19
25. Park SH, Lee HS, Choe JH, Lee JS, Seo JT. Success rate of microsurgical multiple testicular sperm extraction and sperm presence in the ejaculate in Korean Men with Y chromosome microdeletions. Korean J Urol. 2013;54(8):536–540. doi:10.4111/kju.2013.54.8.536
26. Bernie AM, Mata DA, Ramasamy R, Schlegel PN. Comparison of microdissection testicular sperm extraction, conventional testicular sperm extraction, and testicular sperm aspiration for nonobstructive azoospermia: a systematic review and meta-analysis. Fertil Steril. 2015;104(5):1099–1103.e3. doi:10.1016/j.fertnstert.2015.07.1136
27. Silber SJ. The Y chromosome in the era of intracytoplasmic sperm injection: a personal review. Fertil Steril. 2011;95(8):2439–2448.e5. doi:10.1016/j.fertnstert.2011.05.070
28. Brandell RA, Mielnik A, Liotta D, et al. AZFb deletions predict the absence of spermatozoa with testicular sperm extraction: preliminary report of a prognostic genetic test. Hum Reprod. 1998;13(10):2812–2815. doi:10.1093/humrep/13.10.2812
29. Stouffs K, Vloeberghs V, Gheldof A, Tournaye H, Seneca S. Are AZFb deletions always incompatible with sperm production? Andrology. 2017;5(4):691–694. doi:10.1111/andr.12350
30. Fernandes S, Huellen K, Goncalves J, et al. High frequency of DAZ1/DAZ2 gene deletions in patients with severe oligozoospermia. Mol Hum Reprod. 2002;8(3):286–298. doi:10.1093/molehr/8.3.286
31. Krausz C, Chianese C, Giachini C, Guarducci E, Laface I, Forti G. The Y chromosome-linked copy number variations and male fertility. J Endocrinol Invest. 2011;34(5):376–382. doi:10.1007/BF03347463
32. Saxena R, Brown LG, Hawkins T, et al. The DAZ gene cluster on the human Y chromosome arose from an autosomal gene that was transposed, repeatedly amplified and pruned. Nat Genet. 1996;14(3):292–299. doi:10.1038/ng1196-292
33. Seboun E, Barbaux S, Bourgeron T, et al. Gene sequence, localization, and evolutionary conservation of DAZLA, a candidate male sterility gene. Genomics. 1997;41(2):227–235. doi:10.1006/geno.1997.4635
34. Yen PH, Chai NN, Salido EC. The human autosomal gene DAZLA: testis specificity and a candidate for male infertility. Hum Mol Genet. 1996;5(12):2013–2017. doi:10.1093/hmg/5.12.2013
35. Silber SJ, Repping S. Transmission of male infertility to future generations: lessons from the Y chromosome. Hum Reprod Update. 2002;8(3):217–229. doi:10.1093/humupd/8.3.217
36. Cariati F, D’Argenio V, Tomaiuolo R. The evolving role of genetic tests in reproductive medicine. J Transl Med. 2019;17. doi:10.1186/s12967-019-2019-8.
37. Peterlin B, Kunej T, Sinkovec J, Gligorievska N, Zorn B. Screening for Y chromosome microdeletions in 226 Slovenian subfertile men. Hum Reprod. 2002;17(1):17–24. doi:10.1093/humrep/17.1.17
38. Liu X, Li Z, Su Z, et al. Novel Y-chromosomal microdeletions associated with non-obstructive azoospermia uncovered by high throughput sequencing of sequence-tagged sites (STSs). Sci Rep. 2016;6. doi:10.1038/srep21831
39. Poznik GD, Henn BM, Yee M-C, et al. Sequencing Y chromosomes resolves discrepancy in time to common ancestor of males versus females. Science. 2013;341(6145):562–565. doi:10.1126/science.1237619
40. Krausz C, Degl’Innocenti S, Nuti F, et al. Natural transmission of USP9Y gene mutations: a new perspective on the role of AZFa genes in male fertility. Hum Mol Genet. 2006;15(18):2673–2681. doi:10.1093/hmg/ddl198
41. Kleiman SE, Yogev L, Lehavi O, et al. The likelihood of finding mature sperm cells in men with AZFb or AZFb-c deletions: six new cases and a review of the literature (1994–2010). Fertil Steril. 2011;95(6):2005–2012.e4. doi:10.1016/j.fertnstert.2011.01.162
42. Repping S, Skaletsky H, Lange J, et al. Recombination between palindromes P5 and P1 on the human Y chromosome causes massive deletions and spermatogenic failure. Am J Hum Genet. 2002;71(4):906–922. doi:10.1086/342928
43. Jaruzelska J, Korcz A, Wojda A, et al. Mosaicism for 45,X cell line may accentuate the severity of spermatogenic defects in men with AZFc deletion. J Med Genet. 2001;38(11):798–802. doi:10.1136/jmg.38.11.798
44. Suganthi R, Vijesh VV, Vandana N, Fathima Ali Benazir J. Y chromosomal microdeletion screening in the workup of male infertility and its current status in India. Int J Fertil Steril. 2014;7(4):253–266.
45. Dada R, Kumar R, Shamsi MB, et al. Higher frequency of Yq microdeletions in sperm DNA as compared to DNA isolated from blood. Asian J Androl. 2007;9(5):720–722. doi:10.1111/j.1745-7262.2007.00274.x
46. Sakthivel PJ, Swaminathan M. Y chromosome microdeletions in sperm DNA of infertile patients from Tamil Nadu, south India. Indian J Urol. 2008;24(4):480–485. doi:10.4103/0970-1591.44252
47. Le Bourhis C, Siffroi JP, McElreavey K, Dadoune JP. Y chromosome microdeletions and germinal mosaicism in infertile males. Mol Hum Reprod. 2000;6(8):688–693. doi:10.1093/molehr/6.8.688
48. Levine H, Jørgensen N, Martino-Andrade A, et al. Temporal trends in sperm count: a systematic review and meta-regression analysis. Hum Reprod Update. 2017;23(6):646–659. doi:10.1093/humupd/dmx022
49. Sengupta P, Dutta S, Krajewska-Kulak E. The disappearing sperms: analysis of reports published between 1980 and 2015. Am J Mens Health. 2017;11(4):1279–1304. doi:10.1177/1557988316643383
50. Johnson M, Raheem A, De Luca F, et al. An analysis of the frequency of Y-chromosome microdeletions and the determination of a threshold sperm concentration for genetic testing in infertile men. BJU Int. 2019;123(2):367–372. doi:10.1111/bju.14521
51. Liu JL, Peña V, Fletcher SA, Kohn TP. Genetic testing in male infertility – reassessing screening thresholds. Curr Opin Urol. 2020;30(3):317–323. doi:10.1097/MOU.0000000000000764
52. Wu AK, Odisho AY, Washington SL, Katz PP, Smith JF. Out-of-pocket fertility patient expense: data from a multicenter prospective infertility Cohort. J Urol. 2014;191(2):427–432. doi:10.1016/j.juro.2013.08.083
53. Ortac M, Ergul R, Gurcan M, et al. Indication for Y chromosome microdeletion analysis in infertile men: is a new sperm concentration threshold needed? Urology. 2020;146:113–117. doi:10.1016/j.urology.2020.09.032
54. Lin Y-W, Hsu -LC-L, Kuo P-L, et al. Partial duplication at AZFc on the Y chromosome is a risk factor for impaired spermatogenesis in Han Chinese in Taiwan. Hum Mutat. 2007;28(5):486–494. doi:10.1002/humu.20473
55. Zhou R, Cheng J, Ma D, et al. Identifying novel copy number variants in azoospermia factor regions and evaluating their effects on spermatogenic impairment. Front Genet. 2019;10:427. doi:10.3389/fgene.2019.00427
56. Repping S, Skaletsky H, Brown L, et al. Polymorphism for a 1.6-Mb deletion of the human Y chromosome persists through balance between recurrent mutation and haploid selection. Nat Genet. 2003;35(3):247–251. doi:10.1038/ng1250
57. de Llanos M, Ballescà JL, Gázquez C, Margarit E, Oliva R. High frequency of gr/gr chromosome Y deletions in consecutive oligospermic ICSI candidates. Hum Reprod. 2005;20(1):216–220. doi:10.1093/humrep/deh582
58. Ferlin A, Tessari A, Ganz F, et al. Association of partial AZFc region deletions with spermatogenic impairment and male infertility. J Med Genet. 2005;42(3):209–213. doi:10.1136/jmg.2004.025833
59. Giachini C, Laface I, Guarducci E, Balercia G, Forti G, Krausz C. Partial AZFc deletions and duplications: clinical correlates in the Italian population. Hum Genet. 2008;124(4):399–410. doi:10.1007/s00439-008-0561-1
60. Rozen SG, Marszalek JD, Irenze K, et al. AZFc deletions and spermatogenic failure: a population-based survey of 20,000 Y chromosomes. Am J Hum Genet. 2012;91(5):890–896. doi:10.1016/j.ajhg.2012.09.003
61. Levkova M, Chervenkov T, Angelova L. The association of gr/gr deletion in the Y chromosome and impaired spermatogenesis in Bulgarian males: a pilot study. Middle East Fertil Soc J. 2020;25(1):10. doi:10.1186/s43043-020-00020-9
62. Hucklenbroich K, Gromoll J, Heinrich M, Hohoff C, Nieschlag E, Simoni M. Partial deletions in the AZFc region of the Y chromosome occur in men with impaired as well as normal spermatogenesis. Hum Reprod. 2005;20(1):191–197. doi:10.1093/humrep/deh558
63. Xie S, Zhang Y, Yang Y. Is the primary AZFc duplication a potential risk for male infertility?: A systematic review and meta-analysis. Andrology. 2020;8(5):996–1004. doi:10.1111/andr.12800
64. Zhang F, Lu C, Li Z, et al. Partial deletions are associated with an increased risk of complete deletion in AZFc: a new insight into the role of partial AZFc deletions in male infertility. J Med Genet. 2007;44(7):437–444. doi:10.1136/jmg.2007.049056
65. Moreno-Mendoza D, Casamonti E, Paoli D, et al. gr/gr deletion predisposes to testicular germ cell tumour independently from altered spermatogenesis: results from the largest European study. Eur J Hum Genet. 2019;27(10):1578–1588. doi:10.1038/s41431-019-0420-7
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.