Back to Journals » Cancer Management and Research » Volume 13
XPG is Modulated by miR-4715-3p and rs873601 Genotypes in Lung Cancer
Authors Yu W, Yao J, Lyu P, Zhou J, Chen X, Liu X, Xiao S
Received 21 December 2020
Accepted for publication 12 March 2021
Published 19 April 2021 Volume 2021:13 Pages 3417—3427
DOI https://doi.org/10.2147/CMAR.S294365
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Eileen O'Reilly
WeiLing Yu,1 JinJian Yao,2 Pengfei Lyu,3 Jing Zhou,4 Xiaoxi Chen,4 Xiaoran Liu,5 Sha Xiao4
1Oncology Department, Haikou City People’s Hospital, Haikou, 570208, Hainan, People’s Republic of China; 2Emergency Department, Hainan General Hospital Affiliated to Hainan Medical University, Haikou, 570311, Hainan, People’s Republic of China; 3Department of Breast-Thoracic Tumor Surgery, The First Affiliated Hospital of Hainan Medical University, Haikou, 570102, Hainan, People’s Republic of China; 4Department of Environmental and Occupational Health, School of Public Health, Hainan Medical University, Haikou, 571199, Hainan, People’s Republic of China; 5Emergency Department, The First Affiliated Hospital of Hainan Medical University, Haikou, Hainan, People’s Republic of China
Correspondence: Xiaoran Liu
Emergency Department, The First Affiliated Hospital of Hainan Medical University, Longhua District, Longhua Road No.31, Haikou, Hainan, 570102, People’s Republic of China
Tel +86-13118901829
Email [email protected]
Sha Xiao
Department of Environmental and Occupational Health, School of Public Health, Hainan Medical University, Longhua District, Xueyuan Road No.3, Haikou, 571199, People’s Republic of China
Tel +86-18808977719
Email [email protected]
Objective: XPG (Xeroderma pigmentosum group G, XPG), a single strand-specific DNA endonuclease in the nucleotide excision repair pathway, has been implicated in lung cancer. Potentially functional rs873601 in XPG is consistently associated with gastrointestinal cancer, and miR-4715-3p, targeting 3UTR of XPG, also influences the process of gastrointestinal carcinogenesis, however, the relationships between XPG and miR-4715-3p and rs873601 in lung cancer have not been elucidated.
Methods: A case-control study included 264 lung cancer patients and 264 cancer-free healthy controls and was designed to determine the relationships between rs873601 and lung cancer and the effect of miR-4715-3p on XPG expression in lung cancer. Fifty matched cases and controls were randomly selected from the lung cancer and control groups to assess the relationships between the expression levels of miR-4715-3p and XPG determined by using qRT-PCR. The association of rs873601 with lung cancer was analyzed by mass spectrometry, and function prediciton of rs873601 genotypes explored by web-based bioinformatics.
Results: miR-4715-3p in the lung cancer group was significantly increased compared with that in the control group (P = 0.011), upregulation of miR-4715-3p correlated with an increase in XPG mRNA (r = 0.399, P < 0.05) in the lung cancer group. The AA genotype was associated with increased risk of lung cancer compared with the AG and GG genotypes of rs873601 (AG vs AA: OR = 0.231, 95% CI: 0.155– 0.345, P < 0.001 GG vs AA: OR = 0.300, 95% CI: 0.131– 0.719, P = 0.003). The genetic association remained significant after adjustment for age, sex, smoking, and drinking, and rs873601-AA was associated with an increase in XPG mRNA in the lung cancer group. The results of web-based bioinformatics analysis indicated rs873601 genotypes might change XPG-RNA stability and bindability between XPG and miR-4715-3p.
Conclusion: Our data characterized that miR-4715-3p and rs873601 genotypes modified XPG expression in lung cancer. These findings may help to elucidate the mechanisms governing lung cancer.
Keywords: miR-4715-3p, XPG, rs873601, lung cancer
Introduction
Lung cancer is the leading malignant tumor with high incidence and mortality, accounting for an estimated 2.2 million new cases and 1.8 million deaths (18%) in 2020.1 Environmental factors, including smoking, drinking, age, air pollution, and lifestyle, have been identified to modulate the risk for lung cancer, and cigarette smoking is a well-recognized risk factor that accounts for approximately 80% of lung cancer cases. Cigarette smoking induces DNA lesions,2,3 the nucleotide excision repair (NER) responses maintain genomic DNA stability and integrity disrupted by DNA lesions, and dysfunction of the NER system is involved in carcinogenesis. XPG (accession number: DI095823), an endonuclease bound to the 3ʹ-ends of the NER system, specifically functions to recognize and remove damaged DNA strands.4 The DNA repair capacity of XPG is related to inherited factors,5 thus, polymorphisms in the NER pathway modify DNA repair capacity.6 XPG has been proposed as a candidate marker for cancer susceptibility, prognosis, and treatment response, including susceptibility to colorectal cancer7 and gastric cancer,8 sensitivity to anticancer treatment of non-small-cell lung cancer (NSCLC),9 and prognosis of advanced NSCLC.10 The rs873601 is the target site of miR-4715-3p binding to XPG mRNA, and the association of rs873601 of XPG with incidence of lung cancer has not been investigated in detail.
MicroRNAs (miRNAs) regulate gene expression through inactivation of complementary sequences in the 3ʹ-untranslated regions (3UTRs) of mRNAs that mediate biological processes in cancer cells.11–13 Bioinformatics analysis identified a miR-4715-3p binding site in the 3UTR of XPG (Supplementary Figure 1), and the target site of miR-4715-3p has been shown to modulate its function in upper gastrointestinal cancer by targeting the 3UTR of 3UTR of aurora kinase A (AURKA).14 The epigenetic interaction between miR-4715-3p and XPG mRNA in lung cancer is poorly understood.
Our study aimed to explore the association of rs873601 with lung cancer and assess the expression of XPG mRNA and miR-4715-3p and the relationship of miR-4715-3p with XPG mRNA in lung cancer to demonstrate potential mechanisms underlying carcinogenesis in lung cancer.
Materials and Methods
Study Population
A total of 264 patients with lung cancer were recruited from Hainan General Hospital and the First Affiliated Hospital of Hainan Medical University from October 2014 to December 2017 based on confirmed pathology. The clinical features of lung cancer were acquired from the medical records of the patients. The clinical data were analyzed according to the eighth edition of the lung cancer stage classification.15 In the case group, 264 subjects were histologically diagnosed with lung cancer, including 185 males and 79 females, with a median age of 62 years. In the control group, 264 healthy controls were recruited during routine physical examination of the population within the same time period, and healthy controls with evidence of malignancy were excluded; 184 subjects were male, and 80 subjects were female, with a median age of 65 years. Informed consent was obtained from the patients, and a questionnaire was used to record the demographic characteristics and medical history, family history, smoking, and drinking; smoking was defined as at least one cigarette per day for more than six months, and drinking was defined as consumption of 50 g of alcohol for more than half a year. Blood samples were collected from peripheral blood using a vacuum blood collection tube with sodium citrate as an anticoagulant, the samples were used for the extraction of miRNA, mRNA, and genomic DNA.
The study was approved by the ethics committee of Hainan Medical University in accordance with the Helsinki Declaration (HYLL-2019-034). Written informed consent was obtained from all patients.
miRNA and mRNA Extraction and Expression Analysis
A whole blood miRNA extraction and purification kit (Beijing Quanshijin Biotechnology Co., Ltd, Beijing, China) was used to extract miRNA. Total RNA was extracted from peripheral blood samples using a column-type whole blood total RNA extraction and purification kit (Shanghai Biotech Biotechnology Co., Ltd, Beijing, China), and the RNA concentration and OD260/OD280 ratio were assessed by a BioPhotometer-Plus nucleic acid and protein analyzer. The cDNA was synthesized using a reverse transcription kit (Beijing Quanshijin Biotechnology Co., Ltd, Beijing, China). The expression levels of XPG mRNA and miR-4715-3p were detected by an SYBR Premix Ex TaqTM II kit (Beijing Quanshijin Biotechnology Co., Ltd, Beijing, China), and human β-actin and U6 genes were used as internal reference genes for relative quantitative analysis.
XPG primers: forward 5ʹ- GGA AGG GAA GAT CCT GGC TGT-3ʹ, reverse 5ʹ- AAG AGT TTG CAG AGC CGA TGA AA-3ʹ; β-actin gene primes: forward 5ʹ- GTC CAC CTT CCA GCA GAT GTG-3ʹ, reverse 5ʹ- GCA TTT GCG GTG GAC GAT-3ʹ. miR-4715-3p and U6 gene primers were purchased from Guangzhou Ruibo Biological Co., Ltd.
The PCR mixture contained 10 μL of SYBR Premix Ex TaqTM (2×), 0.8 μL of PCR forward primer (10 μM), 0.8 μL of PCR reverse primer (10 μM), 0.4 μL of ROX reference dye (50×), 6.0 μL of dH2O, and 2 μL of DNA. The total reaction volume was 20 μL. All samples were run for 40 cycles of 5 s at 95°C and 34 s at 60°C. Each run contained three negative controls with 2 μL of distilled water instead of DNA. All experiments were performed at least in triplicate. The expression levels of XPG mRNA and miR-4715-3p were analyzed using the 2−ΔCt method. ΔCT = CT(target gene) - CT(internal reference genes).
DNA Extraction and SNP Genotyping
A whole blood DNA kit (TaKaRa Company, Dalian, China) was used to extract genomic DNA from the blood sample, and a BioPhotometer-Plus nucleic acid and protein analyzer was used to measure the DNA concentration and OD260/OD280 ratio. The polymorphism of XPG rs873601 was detected by mass spectrometry.
Statistical Methods
Epidata was used to store the questionnaire data, and SPSS 25.0 was used for statistical analysis. Nonparametric tests of two independent samples were used to compare the relative expression levels of miR-4715-3p and XPG mRNA between the lung cancer and control groups, and the Spearman rank correlation was used to assess the correlation of miR-4715-3p and XPG mRNA expression in the lung cancer group. The genotypic and allelic distribution of rs873601 between the lung cancer and control groups was evaluated by the chi-squared test, and the odds ratio (OR) and 95% confidence interval (CI) were calculated by SPSS 25.0. A P value less than 0.05 was considered significant.
Function Exploration Based on Bioinformatics Database
The sequence interaction exploration and prediction by RNAfold web-based bioinformatics, including RNAfold (http://rna.tbi.univie.ac.at//cgi-bin/RNAWebSuite/RNAfold.cgi), TargetScan (http://www.targetscan.org/vert_71/) and mirSNP (http://bioinfo.bjmu.edu.cn/mirsnp/search/). Those web-based database predict the biological effect of the significant SNPs and miR-4715-3p on XPG. RNAfold is a database for prediction of RNA structure and energy changes in RNA folding based on the sequence. Free energy represents the energy required to change the secondary RNA structure. MirSNP is available online database that collect human SNPs in predicted miRNA-mRNA binding sites whether an SNP within the target site would decrease/break or enhance/create miRNA-mRNA binding site.
Results
Characteristics of Enrolled Subjects
The characteristics of drinking, smoking, age, and sex in the lung cancer (N=264) and control groups (N=264) were not significantly different (Table 1). Fifty matched subjects at a ratio of 1:1 were randomly selected from the two groups to compare XPG mRNA and miR-4715-3p levels between the groups. No significant differences in drinking, smoking, age, or sex were observed in these two matched groups.
 |
Table 1 Characteristics of 528 Subjects Enrolled in the Study |
Expression of XPG mRNA and miR-4715-3p in the Lung Cancer and Control Groups
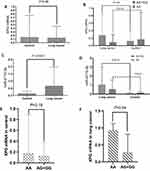
The levels of circulating XPG mRNA and miR-4715-3p in the two groups were not normally distributed; therefore, the nonparametric rank sum test was used to evaluate the differences. XPG mRNA was not significantly different between the lung cancer and control groups (Figure 1A and B, Table 2) and in the genotype groups (Figure 1B). The serum level of miR-4715-3p in the lung cancer group was significantly increased compared with that in the control group (Figure 1C, Table 2), and the genotypes of rs873601 contributed to the significance of differences in miR-4715-3p (Figure 1D).
 |
Table 2 Expression of XPG mRNA and miRNA-4715-3p |
The expression levels of miR-4715-3p and XPG mRNAs were used for stratification into a high group and a low group based on the median expression value in the control group. No significant differences in XPG mRNA between the control and lung cancer groups were observed (Table 3), and patients with high serum levels of miR-4715-3p had an increased risk of lung cancer compared to that in patients with low expression of miR-4715-3p (Table 3).
 |
Table 3 The Relationships Between the Expression of miRNA-4715-3p and XPG mRNA |
Expression of miR-4715-3p and XPG mRNA in 50 Lung Cancer Cases
The expression levels of circulating miR-4715-3p and XPG mRNA in the lung cancer group were stratified by covariates (alcohol, smoking, age, sex, and rs873601), there were no significant differences in the plasma levels of miR-4715-3p. The relative expression level of XPG mRNA in patients older than 65 years of age was significantly higher than that in patients younger than 65 years of age. The level of XPG mRNA was significantly increased in patients with rs873601-AA compared with that in patients with the rs873601-AG+GG genotype in the lung cancer group (Figure 1E and F, Table 4). We detected a correlation between upregulation of miR-4715-3p and increase of XPG mRNA (r = 0.399, P <0.05) in the lung cancer group.
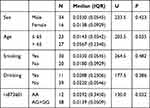 |
Table 4 Expression of XPG mRNA in Lung Cancer |
XPG rs873601 Susceptibility to Lung Cancer
Three genotypes of rs873601 included wild-type (AA), heterozygous (AG), and mutant homozygous (GG). The frequencies of the three genotypes were 48.5%, 47.0%, and 4.5% in the lung cancer group and 18.2%, 76.1%, and 5.7% in the control group, respectively. Significant differences in the genotype distribution between the lung cancer group (N=264) and control group (N=264) were detected (χ2 = 66.398, P <0.001). Patients with the AG or GG genotypes had a decreased risk of lung cancer compared with that of patients carrying the AA genotype. Patients with at least one G allele had a decreased risk. Adjustment for age, sex, smoking, and drinking, XPG rs873601 was still associated with lung cancer (Table 5).
 |
Table 5 Association of XPG rs873601 with Lung Cancer |
Association of XPG rs873601 with Lung Cancer Stratified by Age
Age stratification indicated that the risk for lung cancer in patients with the AG (OR=0.308, 95% CI: 0.15–0.62) and AG + GG (OR=0.32, 95% CI: 0.16–0.63) genotypes was reduced at ages less than 55 years compared with that for the AA genotype. In the 55–65 age group, the risk for lung cancer was also decreased in patients with the AG (OR=0.15, 95% CI: 0.06–0.38), GG (OR=0.04, 95% CI: 0.00–0.38), and AG + GG (OR=0.14, 95% CI: 0.06–0.35) genotypes. In the 65–75 age group, the risk for lung cancer in patients with the AG (OR = 0.29, 95% CI: 0.14–0.60) and AG + GG (OR = 0.30, 95% CI: 0.15–0.62) genotypes was also decreased. In the age group older than 75 years, the lung cancer risk in patients with the AG and AG + GG genotypes was reduced (OR = 0.14, 95% CI: 0.05–0.41; OR = 0.16, 95% CI: 0.057–0.454, respectively) compared with that for the AA genotype (Table 4). Adjustment for sex, alcohol, and smoking, the risk for lung cancer was reduced in patients with the AG, GG, and AG + GG genotypes (ORadj = 0.23, 95% CI: 0.16–0.35; corrected ORadj = 0.30, 95% CI: 0.129–0.682; ORadj = 0.237, 95% CI: 0.159–0.352, respectively) compared to patients with AA genotype (Table 6).
 |
Table 6 The Association of XPG rs873601 with Lung Cancer Stratified by Age |
Association of XPG rs873601 with Lung Cancer Stratified by Gender
In the female group, the lung cancer risk for the AG and AG + GG genotypes was reduced (OR = 0.24, 95% CI: 0.12–0.48; OR = 0.24, 95% CI: 0.12–0.48, respectively) compared with that for the AA genotype. In male patients, the lung cancer risk for the AG, GG, and AG + GG genotypes was reduced compared with that for the AA genotype (OR = 0.23, 95% CI: 0.14–0.37; OR = 0.31, 95% CI: 0.12–0.78; OR=0.24, 95% CI: 0.15–0.38, respectively). Adjustment for age, alcohol, and smoking, the patients with AG (ORadj = 0.23, 95% CI: 0.15–0.34), GG (ORadj = 0.29, 95% CI: 0.13–0.67), and AG + GG (ORadj = 0.23, 95% CI: 0.16–0.36) genotypes had considerably lower risk of lung cancer compared to patients with the AA genotype (Table 7).
 |
Table 7 Association of XPG rs873601 with Lung Cancer Stratified by Sex |
Association of XPG rs873601 with Lung Cancer Stratified by Smoking
In the smoking group, the lung cancer risk for the AG, GG, and AG + GG genotypes was decreased compared with that for the AA genotype (OR = 0.29, 95% CI: 0.17–0.50; OR = 0.20, 95% CI: 0.07–0.60; OR = 0.28, 95% CI: 0.17–0.48, respectively). In the non-smoking group, the AG and AG + GG genotypes reduced the risk of lung cancer compared with that for the AA genotype (OR = 0.18, 95% CI: 0.10–0.32; OR = 0.19, 95% CI: 0.11–0.35, respectively). Adjustment for age, sex, and drinking indicated that the risk for lung cancer in patients with the AG (ORadj = 0.23, 95% CI: 0.16–0.35), GG (ORadj = 0.30, 95% CI: 0.13–0.69), and AG + GG (ORadj = 0.24, 95% CI: 0.16–0.35) genotypes was decreased compared to that for the AA genotype (Table 8).
 |
Table 8 Association of XPG rs873601 with Lung Cancer Stratified by Smoking |
Association XPG rs873601 with Lung Cancer Stratified by Drinking
In the drinking stratification, the distribution of the subjects between the AA genotype and AG and AG + GG genotypes was significantly different. The lung cancer risk for the AG and AG + GG genotypes was reduced compared with that for the AA genotype (OR = 0.24, 95% CI: 0.10–0.55; OR = 0.24, 95% CI: 0.11–0.56, respectively). In the non-drinking group, the AG (OR = 0.23, 95% CI: 0.14–0.36), GG (OR = 0.31, 95% CI: 0.13 ~ 0.73), and AG + GG (OR = 0.23, 95% CI: 0.15–0.37) genotypes decreased the risk for lung cancer compared with that for the AA genotype. The AG (ORadj = 0.23, 95% CI: 0.15–0.34), GG (ORadj =0.29, 95% CI: 0.13–0.67), and AG + GG (ORadj =0.23, 95% CI: 0.16–0.35) genotypes also decreased the risk for lung cancer adjusted for age, sex, and smoking status compared with that for the AA genotype (Table 9).
 |
Table 9 Association of XPG rs873601 and Lung Cancer Stratified by Drinking |
Bioinformatics Analysis Predicted Structural Differences Corresponding to the rs873601 Genotypes
RNAfold analysis of the effect of rs873601 on the 3UTR of XPG used a 105 bp (rs873601 at 91st) sequence of the 3UTR of XPG to predict the changes in the secondary structure corresponding to the rs873601 genotypes [Figure 2]. The secondary structures corresponding to rs873601 are shown in Figure 3 (centroid secondary and minimum free energy, MFE). The rs873601 genotypes also influenced centroid secondary and MFE of the thermodynamic ensemble, and MFE of the thermodynamic ensemble was changed from −17.63kcal/mol (rs873601-A) to −15.73kcal/mol (rs873601-G), indicating that rs873601 may change the structural stability and miRNA binding ability of the 3UTR of XPG, and the rs873601 also change binding process of miR-4715-3p based on mirSNP prediction (Supplementary Figure 3).
 |
Figure 2 The RNAfold algorithm predicts the genotypic impact of rs873601 on sequence structure of XPG. (A) The sequence structure of rs873601-A. (B) The sequence structure of rs873601-G. |
Discussion
The present study characterized the impact of miR-4715-3p and rs873601 on XPG mRNA. The level of miR-4715-3p was significantly increased, and a genotypic difference between lung cancer and the control group was detected; additionally, miR-4715-3p was positively associated with XPG mRNA. The rs873601-AA polymorphism demonstrated susceptibility to lung cancer after adjustment and stratification. The genotypic variants of rs873601 were functionally associated with differential expression of XPG mRNA in lung cancer.
The XPG gene, locating at the chromosomal locus 13q33, consists of 15 exons, XPG corrects the excision in multiple DNA damage responses. The correction by XPG is regulated by sequences and motifs that scaffold and sculpture damaged DNA by binding both NER bubble junctions and replication forks.4,16 Genetic variants and posttranscriptional regulation alter functional motifs, and alterations in functional motifs may reduce DNA repair capacity and lead to increased cancer risks.17,18 Our results revealed that miR-4715-3p modulated XPG expression and may have altered its repair ability, resulting in lung cancer [Supplementary Figure 2].
Significant upregulation of miR-4715-3p was observed in the lung cancer group compared with that in the control group, genotypic analysis of miR-4715-3p expression indicated that miR-4715-3p upregulation was equally significant in both lung cancer and control groups. Notably, increase of miR-4715-3p correlated with upregulation of XPG mRNA in the lung cancer group. A recent study detected downregulation of miR-4715-3p in upper gastrointestinal adenocarcinoma (UCG), miR-4715-3p has putative binding sites in the 3UTR of AURKA, and miR-4715-3p mediated a reduction in AURKA levels, which in turn resulted in GPX4 inhibition and cell ferroptosis.4 Bioinformatics analysis using TargetScan indicated that miR-4715-3p targeted the 7mer-m8 site in the 3UTR of XPG [Supplementary Figure 2], and the target site covered the rs873601 region, partially contributing to interaction between rs873601 and miR-4715-3p.
MiR-4715-3p might indirectly associate with an increase in XPG unlike the model suggesting direct regulation of AURKA by miR-4715-3p. This discrepancy may be due to the differences in cancer types, regulatory pathways or polymorphism of miR-4715-3p target site, or miR-4715-3p may also play dual roles in upregulation and downregulation of the expression of various target genes.13 Additionally, lung cancer-associated transcript 1 (RAC1) is an oncogenic regulator that promotes cell proliferation and metastasis, and miR-4715-3p sponges long noncoding RNA LCAT1, which functions as a ceRNA to regulate RAC1 in lung cancer,19 therefore, an increase in miR-4715-3p levels associated with upregulation of the expression of XPG mRNA may link miR-4715-3p and the NER repair system. The XPG gene functions by stabilizing the structure of DNA and maintaining the DNA damage repair capacity,20 miR-4715-3p may influence the maintenance of genomic integrity by upregulating the expression of XPG.
XPG expression of rs873601-AA genotype confer to an increase in peripheral blood compared with that of the AG + GG genotypes in the lung cancer group. The rs873601-AA genotype is associated with risk for some cancers,7,21 the expression of XPG mRNA was decreased in adjacent non-tumor and normal gastric tissues,22 which in turn indicate rs873601-AA demonstrated the risk factor in lung cancer. Our findings indicated that a genotype-dependent change in XPG mRNA may have implications in lung carcinogenesis. Indeed, bioinformatics prediction indicated that rs873601 was located in the 3UTR corresponding to the miR-4715-3p binding site of XPG mRNA (Supplementary Figure 1), and polymorphisms in these regions can alter mRNA expression and miRNA-mRNA binding (Supplementary Figure 2), SNPs in the miRNA genes and miRNA target genes have been proposed to influence cancer risk by altering the expression levels of target miRNAs.23
Our analysis demonstrated that rs873601 may influence the secondary structure of the 3UTR of XPG mRNA to a certain extent, and the differences in the structure induced by the genotypes were in part related to an increase in the circulating level of XPG and its involvement in lung carcinogenesis. An age over 65 years was also related to increased expression of XPG mRNA in the lung cancer group, indicating that increased XPG expression may be associated with increased age.
Previous studies reported that rs873601-AA was associated with some cancers, and XPG rs873601 AA increased the risk of glioma,24 uterine leiomyoma,25 and gastric cancer.26 Our results extend susceptibility of rs873601 to lung cancer, however, rs873601 is not associated with NSCLC that may be due to the fact that most patients enrolled in the study were advanced cases at stage IV (67.4%) and their histological type concentrated on adenocarcinoma (73.2%).10 Notably, the XPG Asp1104His (rs17655) polymorphism is related to the risk for lung cancer in African Americans27 and for colorectal cancer in Asians,28 the HapMap analysis indicated that rs17655, a well-defined SNP, is in linkage disequilibrium with rs873601 (R2=0.91), suggesting that the functions and susceptibility have identical as rs873601 for cancer genetics. MirSNP predicts that rs873601-A enhances the binding activity of miR-4715-3p, and rs873601-A decrease the binding activity of miR-4715-3p29 (Supplementary Figure 3), the polymorphism of target site among in target gene, miRNA or lncRNA could potentially alter stability of binding.30,31 Together with these, our results confirmed that the rs873601 polymorphism of XPG is associated with susceptibility and biological implications to lung cancer.
However, the study has several limitations, including comparatively small sample size used for analysis; all subjects were from southern China, which may influence statistical power. The application of the results for other populations requires caution because the population of the present study was restricted to unrelated Chinese Han ethnicity. Finally, the tumor types, stages, pathological types, and prognosis were not assessed.
The results indicated that XPG is modulated by miR-4715-3p and rs873601, suggesting that miR-4715-3p and rs873601 influence XPG expression, altering DNA repair capacity in lung cancer. Rs873601 locate at miR-4715-3p-binding site in XPG mRNA; thus, XPG expression may alter the miRNA–mRNA interaction due to genotypic differences. These molecular and functional studies require further validation and exploration.
Acknowledgments
Weiling Yu and Jinjian Yao are co-first authors in this manuscript. This study was supported by the National Natural Science Foundation of China (No 81660550), the Province Natural Science Foundation of Hainan (No 814292 and No 310150), and the Foundation for Fostering Talents of Hainan Medical University (No HYPY201911).
Disclosure
The authors state that they have no conflicts of interest to disclose.
References
1. Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021:1–41. doi:10.3322/caac.21660
2. Hirao T, Nelson HH, Ashok TDS, et al. Tobacco smoke-induced DNA damage and an early age of smoking initiation induce chromosome loss at 3p21 in lung cancer. Cancer Res. 2001;61(2):612–615.
3. Subramanian J, Govindan R. Lung cancer in never smokers: a review. J Clin Oncol. 2007;25(5):561–570. doi:10.1200/JCO.2006.06.8015
4. Tsutakawa SE, Sarker AH, Ng C, et al. Human XPG nuclease structure, assembly, and activities with insights for neurodegeneration and cancer from pathogenic mutations. PNAS. 2020;117(25):14127–14138. doi:10.1073/pnas.1921311117
5. Li C, Hu Z, Liu Z, et al. XPG gene polymorphisms and cancer susceptibility: evidence from 47 studies. Cancer Epidemiol Biomarkers Prev. 2006;15(12):2526–2532. doi:10.1158/1055-9965.EPI-06-0672
6. Li C, Yu X, Guo D, et al. Association between common polymorphisms in ERCC gene and prognosis of osteosarcoma in patients treated with chemotherapy: a meta-analysis. Onco Targets Ther. 2018;18(11):3495–3504. doi:10.2147/OTT.S158167
7. He J, Qiu LX, Wang MY, et al. Polymorphisms in the XPG gene and risk of gastric cancer in Chinese populations. Hum Genet. 2012;131:1235–1244. doi:10.1007/s00439-012-1152-8
8. Chen Y-Z, Guo F, Sun H-W, et al. Association between XPG polymorphisms and stomach cancer susceptibility in a Chinese population. J Cell Mol Med. 2016;20(5):903–908. doi:10.1111/jcmm.12773
9. Wang C, Nie H, Li Y, et al. The study of the relation of DNA repair pathway genes SNPs and the sensitivity to radiotherapy and chemotherapy of NSCLC. Sci Rep. 2016;6:26526. doi:10.1038/srep26526
10. Yuli Y, Zhe S, Xia WA, et al. XPG is a novel biomarker of clinical outcome in advanced non-small-cell lung cancer. Pak J Med Sci. 2013;29(3):762–767. doi:10.12669/pjms.293.3664
11. Lee S, Vasudevan S. Post-transcriptional stimulation of gene expression by microRNAs. Adv Exp Med Biol. 2013;768:97–126. doi:10.1007/978-1-4614-5107-5_7
12. Giorgio S, Morelli MB, Santoni M, et al. Targeting transient receptor potential channels by MicroRNAs drives tumor development and progression. Adv Exp Med Biol. 2020;1131:605–623. doi:10.1007/978-3-030-12457-1_24
13. Persson H, Kvist A, Rego N, et al. Identification of new MicroRNAs in paired normal and tumor breast tissue suggests a dual role for the ERBB2/Her2 gene. Cancer Res. 2011;71(1):78–86. doi:10.1158/0008-5472.CAN-10-1869
14. Gomaa A, Peng D, Chen Z, et al. Epigenetic regulation of AURKA by miR-4715-3p in upper gastrointestinal cancers. Sci Rep. 2019;9(1):16970. doi:10.1038/s41598-019-53174-6
15. Detterbeck FC, Boffa DJ, Kim AW, et al. The eighth edition lung cancer stage classification. Chest. 2017;151(1):193–203. doi:10.1016/j.chest.2016.10.010
16. Wakasugi M, Reardon JT, Sancar A. The non-catalytic function of XPG protein during dual incision in human nucleotide excision repair. J Biol Chem. 1997;272(25):16030–16034. doi:10.1074/jbc.272.25.16030
17. He J, Wang F, Zhu J, et al. Association of potentially functional variants in the XPG gene with neuroblastoma risk in a Chinese population. J Cell Mol Med. 2016;20(8):1481–1490. doi:10.1111/jcmm.12836
18. Zhang X, Crawford EL, Blomquist TM, et al. Haplotype and diplotype analyses of variation in ERCC5 transcription cis-regulation in normal bronchial epithelial cells. Physiol Genomics. 2016;48(7):537–543. doi:10.1152/physiolgenomics.00021.2015
19. Yang J, Qiu Q, Qian X, et al. Long noncoding RNA LCAT1 functions as aceRNA to regulate RAC1 function by sponging miR-4715-5p in lung cancer. Mol Cancer. 2019;18(1):171. doi:10.1186/s12943-019-1107-y
20. González-Corrochano R, Ruiz FM, Taylor NM, et al. The crystal structure of human XPG, the xeroderma pigmentosum group G endonuclease, provides insight into nucleotide excision DNA repair. Nucleic Acids Res. 2020;48(17):9943–9958. doi:10.1093/nar/gkaa688
21. Han C, Huang X, Hua R, et al. The association between XPG polymorphisms and cancer susceptibility: evidence from observational studies. Medicine. 2017;96(32):7467–7468. doi:10.1097/MD.0000000000007467
22. Deng N, Liu J-W, Sun L-P, et al. Expression of XPG protein in the development, progression and prognosis of gastric cancer. PLoS One. 2014;9(9):e108704. doi:10.1371/journal.pone.0108704
23. Mullany LE, Wolff RK, Herrick JS, Buas MF, Slattery ML. Slattery, SNP regulation of microRNA expression and subsequent colon cancer risk. PLoS One. 2015;10(12):e0143894. doi:10.1371/journal.pone.0143894
24. Yuan L, Hu WM, Chen K, et al. XPG gene polymorphisms and glioma susceptibility: a two-centre case-control study. Br J Biomed Sci. 2021:1–6. doi:10.1080/09674845.2020.1870308
25. Liu Z-Q, Chen G-G, Sun R-L, et al. XPG rs873601 G>A contributes to uterine leiomyoma susceptibility in a Southern Chinese population. Biosci Rep. 2018;38(5):BSR20181116. doi:10.1042/BSR20181116
26. Hua RX, Zhuo ZJ, Zhu J, et al. Association between genetic variants in the XPG gene and gastric cancer risk in a Southern Chinese population. Aging. 2016;8(12):3311–3320. doi:10.18632/aging.101119
27. Chang JS, Wrensch MR, Hansen HM, et al. Nucleotide excision repair genes and risk of lung cancer among San Francisco Bay Area Latinos and African Americans. Int J Cancer. 2008;123(9):2095–2104. doi:10.1002/ijc.23801
28. Su J, Zhu Y, Dai B, et al. XPG Asp1104His polymorphism increases colorectal cancer risk especially in Asians. Am J Transl Res. 2019;11(2):1020–1029.
29. Liu C, Zhang F, Li T, et al. MirSNP, a database of polymorphisms altering miRNA target sites, identifies miRNA-related SNPs in GWAS SNPs and eQTLs. BMC Genomics. 2012;23(13):661. doi:10.1186/1471-2164-13-661
30. Yu WL, Yao JJ, Xie Z, et al. LncRNA PRNCR1 rs1456315 and CCAT2 rs6983267 polymorphisms on 8q24 associated with lung cancer. Int J Gen Med. 2021;14:255–266. doi:10.2147/IJGM.S290997
31. Yan Y-X, Xiao H-B, Zhang J, et al. Pri-miR-144 rs9279 is associated with type 2 diabetes and regulation of stress response. J Cell Physiol. 2021;236(1):561–569. doi:10.1002/jcp.29883
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.