Back to Journals » International Journal of General Medicine » Volume 14
WGCNA Analysis Identifies Polycystic Ovary Syndrome-Associated Circular RNAs That Interact with RNA-Binding Proteins and Sponge miRNAs
Authors Li M, Zeng Z, Zhang A, Ye Q, Su S, Xia T
Received 19 August 2021
Accepted for publication 2 November 2021
Published 23 November 2021 Volume 2021:14 Pages 8737—8751
DOI https://doi.org/10.2147/IJGM.S335108
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Mengxiong Li,1,* Zhi Zeng,2,* Aiqing Zhang,3 Qingjian Ye,4 Shujun Su,4 Tingting Xia5
1Department of Obstetrics and Gynecology, The Seventh Affiliated Hospital of Sun Yat-sen University, Shenzhen, Guangdong, 518107, People’s Republic of China; 2The Department of Reproductive Medicine Center, The Sixth Affiliated Hospital of Sun Yat-sen University, Guangzhou, Guangdong, 510000, People’s Republic of China; 3Department of Obstetrics and Gynecology, The Third Affiliated Hospital of Sun Yat-Sen University, Guangzhou, Guangdong, 510630, People’s Republic of China; 4Department of Gynecology, The Third Affiliated Hospital of Sun Yat-Sen University, Guangzhou, Guangdong, 510630, People’s Republic of China; 5Center for Reproductive Medicine, The Third Affiliated Hospital of Sun Yat-Sen University, Guangzhou, Guangdong, 510630, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Tingting Xia Tel +86-020-85256032
Email [email protected]
Objective: Dysfunction of cumulus granulosa cells has been suggested as a contributor to abnormal folliculogenesis and the development of polycystic ovary syndrome (PCOS), but the underlying molecular mechanisms remain unclear. Recent studies indicate that circular RNAs (circRNAs) exert important roles for diseases. We aimed to screen crucial circRNAs of PCOS patients and predict their functions.
Methods: The high-throughput datasets of circRNAs (GSE145296), microRNAs (miRNAs; GSE72274) and messenger RNAs (mRNAs; GSE155489) in cumulus cells of PCOS patients and controls were collected from the Gene Expression Omnibus database. Differentially expressed circRNAs (DECs), miRNAs (DEMs) and protein-coding genes (DEGs) were identified by the limma method. The weighted correlation network analysis (WGCNA) was conducted using the DECs to mine PCOS-associated modules. Hub DECs in modules were defined as both of |gene significance| and |module membership| > 0.8. The downstream effectors of hub DECs were predicted by constructing DEC-DEM-DEG ceRNA and DEC-RNA binding protein (RBP) networks. Gene Ontology and Kyoto Encyclopedia of Genes and Genomes enrichment analyses were performed to explore the functions of circRNAs.
Results: A total of 3614 DECs, 3544 DEGs and 1469 DEMs were identified between PCOS and controls. WGCNA analysis yielded five PCOS-related modules, of which 190 DECs were hub circRNAs. Seventeen hub DECs, nine DEMs, and 315 DEGs were identified to construct the ceRNA network, while 56 hub DECs and two DEGs (MBNL2, RBPMS) constituted the circRNA-RBP network. Five hub DECs (hsa_circ_0063309, hsa_circ_0054275, hsa_circ_0056196, hsa_circ_0018108 and hsa_circ_0070987) were overlapped between ceRNA and DEC-MBNL2 regulatory networks and thus they may be pivotal for PCOS. Furthermore, hsa_circ_0099109 could interact with the RBP gene RBPMS. Function analyses showed these circRNAs were inflammation-, apoptosis- or steroidogenesis-related.
Conclusion: Aberrant expression of six circRNAs that function as RBP regulators or miRNA sponges may be possible mechanisms underlying the pathogenesis of PCOS by affecting apoptosis and steroidogenesis in cumulus cells.
Keywords: circular RNAs, competitive endogenous RNAs, polycystic ovary syndrome, RNA-binding proteins, weighted gene co-expression network analysis
Introduction
Polycystic ovary syndrome (PCOS) is a common endocrine metabolic disorder in women of childbearing age.1 PCOS patients are typically characterized by androgen excess, polycystic ovaries and anovulation, and are often accompanied by insulin resistance and obesity.2 Consequently, patients with this syndrome not only suffer from infertility,3 but also have an increased risk of developing diabetes, hypertension, cardiovascular diseases and cancer.4 However, management of this condition is still challenging, with anti-androgen and ovulation stimulating hormones as well as insulin-sensitizing drugs predominantly, which may cause serious side effects.2 These findings indicate the necessity to deeply explore the molecular mechanisms of PCOS to develop more effective therapeutic strategies.
Ovarian follicular development and ovulation is a complex process that involves the communication between the oocytes and the surrounding granulosa cells (especially the cumulus subset).5 The cumulus granulosa cells were reported to promote oocyte growth by providing energy source (eg pyruvate, free fatty acids), steroid hormone (conversion of cholesterol to pregnenolone, progesterone, estradiol in turn under the binding of luteinizing hormone receptor and follicle-stimulating hormone with their corresponding ligands) and various growth regulators (eg epidermal growth factor) through gap junctions.5–8 Dysfunction of cumulus granulosa cells (including increased apoptosis, decreased proliferation, estrogen deficiency and androgen accumulation) led to abnormal folliculogenesis and the development of PCOS.9–11 Therefore, investigation of molecular mechanisms associated with dysfunction of cumulus cells may provide important insights into the pathogenesis of PCOS and potential therapeutic targets.
Circular RNAs (circRNAs) are a class of non-coding RNAs that form a covalently closed-loop structure from back-spliced exons, without a canonical 5′-end cap and 3′-end poly (A) tails.12 Mounting evidence indicates that circRNAs participate in various biological processes by functioning as microRNA (miRNA) sponge, interacting with RNA-binding proteins (RBPs) and regulating the expression of their host genes.13,14 These studies reveal circRNAs may represent attractive players for PCOS. This hypothesis has been demonstrated previously. Ma et al identified 286 circRNAs that were differentially expressed between PCOS and non-PCOS groups by microarray analysis and confirmed hsa_circ_0043533 was upregulated, while hsa_circ_0097636 was downregulated by quantitative reverse transcription polymerase chain reaction (qRT-PCR). The expression levels of hsa_circ_0043533 and hsa_circ_0097636 were positively correlated with serum testosterone in PCOS patients.15 Similarly, Che et al16 and Zhang et al17 used the microarray and qRT-PCR techniques to validate four (hsa_circ_0083952, hsa_circ_0082709, hsa_circ_0002425, hsa_circ_0015168) and two (hsa_circ_0001577, hsa_circ_0020093) circRNAs in the granulosa cells may be related to PCOS disease, respectively. Che et al16 also predicted miRNAs that interacted with circRNAs. In the study of Liu et al, overexpression of circPSMC3 was proved to decrease the insulin release and increase the number of granulosa cells in PCOS model mice by sponging miR-296-3p and then regulating PTEN expression.18 Additionally, in vitro studies implied ciRS-12619 and circASPH20 regulated the proliferation and apoptosis of ovarian granulosa cells by targeting the miR-21-programmed cell death 4-reactive oxygen species and miR-375/mitogen-activated protein kinase kinase 6 axis, respectively. However, PCOS-associated circRNAs remain rarely reported.
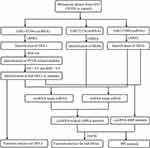
The present study aimed to further screen PCOS-associated circRNAs by re-analysis of the microarray dataset of Ma et al15 based on weighted gene co-expression network analysis (WGCNA)21 that is a powerful systems biology algorithm for finding modules of highly correlated genes and associating the gene modules with clinical features. Furthermore, we also integrated the high-throughput datasets of miRNAs22 and mRNAs in cumulus cells of PCOS patients to construct the circRNA-miRNA-mRNA competing endogenous RNA (ceRNA) network and circRNA-RBP interaction network to predict the function mechanisms of circRNAs (Figure 1). Our study may highlight the importance of crucial circRNAs in the etiology of PCOS. Targeted regulation of these circRNAs may represent underlying treatment strategies for PCOS.
Materials and Methods
Dataset Collection
PCOS-associated expression profile data of GSE145296,15 GSE7227422 and GSE15548923 were obtained from the Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) database by searching with the following keywords: (“circRNA” or “miRNA” or “transcriptomic”) and (“polycystic ovary syndrome”) and (“cumulus cells”). GSE145296 dataset analyzed the circRNAs in cumulus cells from six PCOS patients and six normal control individuals using Agilent-084217 CapitalBio Technology Human CircRNA Array v2. GSE72274 explored the altered miRNA profiles in cumulus cells from five women with PCOS and five without PCOS using Agilent-038166 cbc_human_miR18.0 array. GSE155489 investigated the transcriptome changes of cumulus cells in four women with PCOS compared with four women without PCOS using HiSeq X Ten whole-genome sequencing technology.
Differential Analysis for circRNAs, miRNAs and mRNAs
Differentially expressed circRNAs (DECs), miRNAs (DEMs) and protein-coding genes (DEGs) between PCOS patients and normal controls were identified by using the Bioconductor limma package of R software (v3.34.7; https://bioconductor.org/packages/release/bioc/html/limma.html).24 P-value <0.05 and |log2fold-change (FC)| ≥ 0.5 were set as the statistical thresholds for DECs, DEMs and DEGs. Heatmap was plotted using the “pheatmap” package (v1.0.8; https://cran.r-project.org/web/packages/pheatmap).
Function Enrichment Analysis
To understand the functions of DECs and DEGs, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) (v6.8; http://david.abcc.ncifcrf.gov). P-value <0.05 was considered to be statistically significant.
WGCNA Analysis for Identification of Hub circRNAs for PCOS
The WGCNA R package (v1.61; https://cran.r-project.org/web/packages/WGCNA/index.html)21 was used to construct a weighted correlation network using the expression data profile of DECs. First, Pearson coefficient was calculated to evaluate the correlation between the two genes. To ensure a scale-free topology, the soft threshold power was selected using the pickSoftThreshold function, by which the adjacency matrix was generated. Then, the adjacency matrix was transformed into a topological overlap matrix (TOM). A hierarchical clustering tree was built according to a TOM-based dissimilarity measure. Significant co-expression modules were identified using the DynamicTreeCut function, with the parameters set as minModuleSize ≥ 50 and cutHeight = 0.995. Next, module eigengene (ME, the representation of the overall expression level of each module), gene significance (GS, the correlation of individual genes with clinical trait) and module membership (MM, the correlation of individual genes with ME) were calculated. Pearson’s correlation analysis was performed to explore the correlation between ME and clinical trait as well as between GS and MM, in which p < 0.05 indicated the modules or genes in the modules were significantly associated with PCOS. The hub circRNAs in the significant modules were filtered with the threshold of |GS| > 0.8 and |MM| > 0.8.
Construction of ceRNA and circRNA–RBP Interaction Networks
The circinteractome (https://circinteractome.irp.nia.nih.gov) and starbase (v3.0, http://starbase.sysu.edu.cn) databases were used to predict the miRNAs or RBPs that interacted with DECs. The DEC-DEM pairs predicted by both the circinteractome and starbase databases were suggested to be important, while the DECs-DEGs (RBPs) interaction pairs were selected if they were predicted by any one database (in starbase, CLIP-Data was set as ≥3). The miRwalk database (v2.0; http://www.zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2) that contains 12 prediction programs was utilized to predict the target genes of DEMs. Those DEGs, which were predicted by at least 5 algorithms, were considered as the true candidate target mRNAs of DEMs. Considering that the network would be larger, the DEGs only with adjusted p-value < 0.05 and |log2FC| ≥ 0.5 were used as the target genes of DECs or DEMs. The DEC-DEM and DEM-DEG interaction pairs were used to construct a ceRNA regulatory network. The DEC-RBP interaction pairs were used for establishing a circRNA-RBP network. These two networks were visualized by using Cytoscape (v3.6.1; www.cytoscape.org/).
Construction of a Protein–Protein Interaction (PPI) Network
To screen hub genes and explore their possible function mechanisms, the interactions between DEGs in the ceRNA and circRNA-RBP networks were predicted using the STRING (Search Tool for the Retrieval of Interacting Genes; v10.0; http://stringdb.org/) database. The DEGs with a confidence score ≥0.4 were included to construct the PPI network using Cytoscape. In addition, degree centrality was calculated for each protein in the PPI network using the CytoNCA plugin in Cytoscape software (http://apps.cytoscape.org/apps/cytonca). The hub genes were determined if they possessed a high degree.
Results
Identification of DECs, DEGs and DEMs
A total of 134,751 circRNAs were identified in the GSE145296 dataset. Of them, 3614 circRNAs were differentially expressed in cumulus cells of PCOS patients compared to controls, including 1823 upregulated and 1791 downregulated DECs (Figure 2A). The GSE155489 dataset comprised 22,064 mRNAs, from which 3544 displayed significantly differential expression in cumulus cells of PCOS patients compared to controls, including 1880 upregulated and 1663 downregulated DEGs (Figure 2B). Of 1887 miRNAs in the GSE72274 dataset, 1469 were screened as DEMs, including 35 upregulated and 1434 downregulated (Figure 2C). The crucial DECs, DEGs and DEMs are listed in Table 1.
 |
Table 1 Crucial circRNAs, miRNAs and mRNAs in PCOS |
Function Enrichment Analysis for DECs and DEGs
To reveal the potential functions of identified DECs, the parental genes of circRNAs were annotated, and function enrichment analysis was performed. A total of 215 GO biological process terms (Table S1) and 36 KEGG pathways (Table S2) were enriched, in which DECs were mainly involved in cell proliferation and immune regulation, such as GO:0051301~cell division (TPX2), GO:0000086~G2/M transition of mitotic cell cycle (TPX2), hsa04914:Progesterone-mediated oocyte maturation, hsa04110:Cell cycle and hsa04662:B cell receptor signaling pathway (Figure 3).
 |
Figure 3 Function enrichment analysis for host genes of differentially expressed circRNAs. |
Function enrichment analysis was also performed for all DEGs. The results showed that 80 GO biological process terms (Table S3) and 29 KEGG pathways (Table S4) were enriched. In line with the results of DECs, DEGs were also mainly associated with inflammation-mediated cell apoptosis, such as GO:0042981~regulation of apoptotic process (PEA15, proliferation and apoptosis adaptor protein 15; caspase 2, CASP2), GO:0002223~stimulatory C-type lectin receptor signaling pathway [TAB2, TGF-beta activated kinase 1 binding protein 2; PAK3, p21 (RAC1) activated kinase 3], GO:0045893~positive regulation of transcription, DNA-templated (RUNX2, RUNX family transcription factor 2), hsa05205:Proteoglycans in cancer (FZD4, frizzled class receptor 4), hsa04668:TNF signaling pathway (TAB2) and hsa04660:T cell receptor signaling pathway (PAK3). Furthermore, DEGs also participated in steroidogenesis, including GO:0006695~cholesterol biosynthetic process (HMGCR, 3-hydroxy-3-methylglutaryl-CoA reductase), hsa01130:Biosynthesis of antibiotics (HMGCR) (Figure 4). It was reported that the high androgen accumulation stimulated inflammation and promoted the apoptosis of cumulus cells.25 Therefore, circRNAs that regulate inflammation-, apoptosis- and steroidogenesis-related DEGs may be important for PCOS.
 |
Figure 4 Function enrichment analysis for differentially expressed protein-coding genes. |
WGCNA Analysis Identifies Hub DECs Associated with PCOS
In WGCNA analysis, the soft threshold power was selected as 22 to ensure a scale-free network distribution (scale free index R2 > 0.9; connectivity = 1) (Figure 5A). Five modules were identified by the DynamicTreeCut algorithm (Figure 5B), of which 853, 196, 902, 1547 and 116 DECs were enriched in the blue, brown, grey, turquoise and yellow module, respectively. All these modules were significantly associated with PCOS (Figure 5C). Also, the GS for PCOS and MM in all the modules showed a strongly significant correlation, suggesting genes in these modules were highly correlated with PCOS (Figure 6). Subsequently, based on the threshold of |GS| > 0.8 and |MM| > 0.8, a total of 39, 34, 29, 85 and 3 DECs were screened from the blue, brown, grey, turquoise and yellow module, respectively (Table 1), indicating they may be hub DECs.
Identification of Hub DECs That Function as miRNA and RBP Sponges
To link the DECs and DEGs, we constructed the circRNA-miRNA-mRNA ceRNA and circRNA-RBP regulatory networks. Both the circinteractome and starbase databases predicted 17 hub DECs identified in WGCNA modules could interact with nine DEMs. miRwalk database predicted 315 DEGs were target genes of these nine DEMs. These interaction pairs were used to construct a ceRNA network (Figure 7).
Among all DEGs, only one (RBPMS, RNA binding protein, mRNA processing factor) and four (MBNL2, muscleblind-like splicing regulator 2; YTHDF2, YTH N6-methyladenosine RNA-binding protein 2; DKC1, dyskerin pseudouridine synthase 1; RBM47,-RNA binding motif protein 47) were overlapped with the RBPs in the circinteractome and starbase database, respectively. Thus, these RBPs were selected for predicting the interacted circRNAs. As a result, one upregulated DEC (hsa_circ_0099109) was predicted to bind with RBPMS; 53 DECs were predicted to bind with MBNL2. Thus, these interaction pairs were used to construct a DEC-RBP regulatory network (Figure 8).
Due to the fact that RBPMS and MBNL2 were not enriched in function results, we overlapped the DECs in the above two regulatory networks to indirectly explain their roles in PCOS. The results showed that five DECs were shared (hsa_circ_0018108, hsa_circ_0054275, hsa_circ_0056196, hsa_circ_0063309, hsa_circ_0070987) between two regulatory networks. These findings revealed their related ceRNA and DEC-RBP axes may be particularly pivotal for PCOS, such as hsa_circ_0063309/hsa_circ_0054275/hsa_circ_0056196/hsa_circ_0018108/hsa_circ_0070987-hsa-miR-139-5p-CASP2/ FZD4/HMGCR, hsa_circ_0063309/hsa_circ_0054275/hsa_circ_0056196-hsa-miR-217-FZD4/TAB2/PEA15/RUNX2, hsa_circ_0063309-hsa-miR-346-FZD4/PEA15/RUNX2, hsa_circ_0063309-hsa-miR-421-PAK3/CASP2/RUNX2, hsa_circ_0056196/hsa_circ_0018108-hsa-miR-496-HMGCR/FZD4 and hsa_circ_0018108/hsa_circ_0054275/hsa_circ_0056196/hsa_circ_0063309/hsa_circ_0070987-MBNL2. Also, DEGs in these regulatory axes were demonstrated to be hub genes after analysis of the degree of each protein in the PPI network (PAK3, degree = 13; RUNX2, degree = 9; HMGCR; TAB2, degree = 8; FZD4, degree = 6; CASP2, degree = 5; PEA15, degree = 2) (Figure 9).
 |
Figure 9 Construction of a PPI network for the genes in the circRNA-miRNA-mRNA ceRNA network. Abbreviations: PPI, protein–protein interaction; ceRNA, competing endogenous RNA. |
Discussion
Using WGCNA method, we identified 190 DECs (that were included in five significant modules) associated with the development of PCOS. Six of them (hsa_circ_0063309, hsa_circ_0054275, hsa_circ_0056196, hsa_circ_0018108, hsa_circ_0070987, hsa_circ_0099109) were considered important by integrating with DEMs and DEGs to construct the circRNA-miRNA-mRNA ceRNA and DEC-RBP regulatory networks. Function analysis revealed these circRNAs were mainly involved in inflammation-mediated cell apoptosis and steroidogenesis by influencing the downstream target genes.
Although all these DECs were not reported previously, two of them encoded the same host genes with other circRNAs (hsa_circ_0063309, DDX17; hsa_circ_0070987, ELF2); while these reported circRNAs were demonstrated to play prominent roles in cell proliferation and apoptosis: Li et al found circDDX17 (hsa_circ_0002211) was significantly downregulated in colorectal cancer tissues. Silencing of circDDX17 promoted colorectal cancer cell proliferation and inhibited apoptosis.5,26 Peng et al observed circDDX17 was lowly expressed in breast cancer tissues and cell lines. Overexpression of circDDX17 inhibited cell proliferation and promoted cell apoptosis in breast cancer.27 In vivo experiments by Ren et al showed elevated expression of circDDX17 blocked colorectal cancer cell growth and elevated chemosensitivity of 5-Fu.28 circRNA microarray and qRT-PCR detected circ-ELF2 (hsa_circRNA_103741) were significantly upregulated in glioma tissues. The collapse of circ-ELF2 led to growth arrest and accelerated cell apoptosis.29 The host genes of our other identified DECs were also revealed to mediate the regulation of hormone synthesis, inflammation or cell apoptosis. Ectopic expression of metastasis associated 1 family member 3 (MTA3, host for hsa_circ_0054275) was reported to promote the cAMP-stimulated progesterone secretion and testosterone production.30,31 Treatment with high concentration of insulin significantly attenuated the level of MTA3 protein inLeydig tumor cells.30 Oroxyloside treatment markedly induced apoptosis in glioma cells through upregulating MTA3.32 Actin-related protein 3 (ACTR3, host for hsa_circ_0056196) was proved to amplify B cell receptor signaling and is required for maximal B cell activation and proliferation.33 Estrogen receptor α-engaged macrophages initiated chronic inflammation by upregulating the expression of kinesin family member 5B (KIF5B, host for hsa_circ_0018108).34 CPSF6 was revealed to be upregulated after HIV-1 infection.35 These findings indicate that our identified circRNAs may be involved in PCOS by changing inflammation, apoptosis and steroidogenesis of cumulus cells.
Many studies have found that circRNAs contain miRNA response element and they could work as endogenous miRNA ‘sponges’ to bind to miRNAs and then exert negative regulation on the expression of downstream genes. circDDX17 was implicated to regulate miR-605-cell cycle-related factors (cyclin dependent kinase 1 and p21),27 miR-346/phospholysine phosphohistidine inorganic pyrophosphate phosphatase,36 miR-31-5p-KN motif and ankyrin repeat domains128 axes. Mechanistically, circ-ELF2 acted as a ceRNA for miR-510-5p to positively modulate the expression of mucin 15 at posttranscriptional level.29 However, their roles remain not well understood. In this study, we predicted five circRNAs (hsa_circ_0063309, hsa_circ_0054275, hsa_circ_0056196, hsa_circ_0018108, hsa_circ_0070987) could sponge hsa-miR-139-5p to upregulate the expression of CASP2, FZD4 and HMGCR; hsa_circ_0063309/hsa_circ_0054275/hsa_circ_0056196 could regulate the expression of FZD4/TAB2/PEA15/RUNX2 by sponging hsa-miR-217; hsa_circ_0063309 also functioned as a ceRNA for hsa-miR-346 and hsa-miR-421 to influence the expression of FZD4/PEA15/RUNX2 and PAK3/CASP2/RUNX2, respectively; hsa_circ_0056196/hsa_circ_0018108 acted as a ceRNA to regulate HMGCR/FZD4 function by sponging hsa-miR-496. In these interaction pairs, only the negative regulatory relationships between some miRNAs and target genes were confirmed by wet experiments, including miR-139-5p-FZD437 and miR-217-RUNX2,38,39 while the circRNA-miRNA and other miRNA-target gene interactions were newly identified. Nevertheless, the target DEGs in the above interaction pairs were revealed to be directly or indirectly related with PCOS by involving in inflammation, proliferation, apoptosis or steroidogenesis of granulosa cells. Zhang et al found RUNX2 was upregulated in cumulus cells from PCOS patients compared with controls. Silencing of RUNX2 promoted estradiol production by enhancing the expression of cytochrome P450 family 11 subfamily A member 1 (CYP11A1) and CYP19A1 which were rate-limiting enzymes for estradiol biosynthesis.11 Compared with women without RUNX2, women with RUNX2 expression had lower number of follicles and peak estradiol levels.40 In granulosa cells that had an upregulated expression of RUNX2 due to knockdown of CREB3/Luman gene, estradiol and progesterone synthesis were decreased, anti-apoptotic protein Bcl-2 was downregulated.41 The FZD member gene FZD3 was upregulated in cumulus cells from PCOS patients. This upregulation showed a tight association with estrogen deficiency. Overexpression of FZD3 compromised estrogen production in human granulosa cells.42 Inhibition of PAK3 prevented the secretion of testosterone, while stimulated oestradiol release.43 Addition of PAK3-specific siRNAs reversed the decreased proliferation rate of granulosa cells induced by slit guidance ligand 2.44 PCOS women possessed higher amount of PED/PEA-15 protein levels than that in controls. This increase was positively associated with high total testosterone.45 Kaur et al identified a 1.86-fold increase in the expression of TAB2 in granulosa cells of PCOS non-insulin resistant patients compared with matched controls and function analysis showed this gene was inflammation-related.46 This pro-inflammatory role of TAB2 was further validated in a previous study that showed TAB2 could activate nuclear factor κ-B to promote the production of interleukin-1.47 mRNA expression for CASP2 was shown to be increased with the onset of apoptosis of granulosa cells.48 HMGCR encodes a rate-limiting enzyme (HMG-CoA reductase) in the cholesterol biosynthetic pathway; while cholesterol is a precursor of androgen production. The upregulation of HMGCR was induced by inflammatory agents.49 The addition of simvastatin reduced ovarian androgen production in PCOS women by blocking the expression of HMGCR and inflammatory mediators.50,51 Although miRNAs were not studied in granulosa cells, the expressions of some miRNAs (miR-421,52 and miR-496,53 and miR-34654) were confirmed to be significantly downregulated in patients or mice with diabetes, a comorbidity of PCOS. Overexpression of miR-139-5p was shown to reduce the blood glucose level and alleviate the apoptosis of liver cells in diabetic mice.55 In vitro experiments observed downregulation of miR-217 could induce human podocyte cells apoptosis.56 In accordance with these studies, we also found all related miRNAs were downregulated and mRNAs were upregulated in cumulus cells of PCOS patients findings. These findings implied the possible functions of circRNAs in PCOS.
CircRNAs can also interact with RBPs. In this study, we predicted that the above five circRNAs (hsa_circ_0063309, hsa_circ_0054275, hsa_circ_0056196, hsa_circ_0018108 and hsa_circ_0070987) could interact with MBNL2; hsa_circ_0099109 could interact with RBPMS to promote their upregulated expression in PCOS. In line with our prediction, Che et al verified that RBPMS was a downstream target of lncRNA HUPCOS which was significantly elevated in granulosa cells to promote the accumulation of testosterone in PCOS patients.57 Downregulation of MBNL2 was shown to enhance the ability of dendritic cells to inhibit virus replication and exert anti-inflammatory roles.58 A significant decrease in the percentage of dendritic cells was found in patients with PCOS compared with control.59 These roles of RBPMS and MBNL2 may also indirectly explain the functions of related circRNAs.
There are some limitations in this study. First, hub circRNAs were only preliminarily identified by analysis of high-throughput datasets with small sample size. Their expression and associations with PCOS patients’ pathophysiological characteristics (estrogen, luteinizing hormone, follicle-stimulating hormone testosterone, insulin resistance index, metaphase II oocytes, maturation rate, embryos, and fertilization rate) should be evaluated using a large number of clinical data collected from our hospital. Second, overexpression and knockdown experiments should be conducted in vitro and in vivo to explore whether these hub circRNAs play roles in inflammation, apoptosis and steroidogenesis of cumulus cells in PCOS and whether targeted regulation of them effectively alleviates the symptoms of PCOS. Third, the downstream mechanisms of circRNAs by functioning as miRNA and RBP sponges should be validated by dual-luciferase reporter gene assay, co-immunoprecipitation, gain and loss of function experiments. Fourth, the DEMs and DEGs that were not linked to circRNAs should also be given special attention to demonstrate their importance in the future.
In conclusion, our present study suggests dysfunction of these six circRNAs may be the underlying mechanism to induce the development of PCOS. These circRNAs may mediate the hormone imbalance and apoptosis of cumulus granulosa cells by interaction with miRNAs (hsa-miR-139-5p, hsa-miR-217, hsa-miR-346, hsa-miR-421, hsa-miR-496) to influence their downstream target genes (CASP2, FZD4, HMGCR, TAB2, PEA15, RUNX2, PAK3) or interaction with RBPs (RBPMS, MBNL2).
Data Sharing Statement
All data were downloaded from Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) under GSE145296, GSE72274 and GSE155489 accession number.
Ethics Approval Statement
Approval from the Ethics Committee of the Third Affiliated Hospital of Sun Yat-Sen University was waived because this study was a statistical analysis of public data and no wet experiments of patients were performed by our authors.
Acknowledgments
This work was financially supported by Medical Research Fund of Guangdong Province (No. A2020575) and CSCO-Pilot Cancer Research Fund (No. Y-2019AZQN-1049).
Disclosure
The authors declare that they have no competing interests.
References
1. Wolf WM, Wattick RA, Kinkade ON, Olfert MD. Geographical prevalence of polycystic ovary syndrome as determined by region and race/ethnicity. Int J Environ Res Public Health. 2018;15(11):2589. doi:10.3390/ijerph15112589
2. Escobar-Morreale HF. Polycystic ovary syndrome: definition, aetiology, diagnosis and treatment. Nat Rev Endocrinol. 2018;14(5):270–284. doi:10.1038/nrendo.2018.24
3. Deshpande PS, Gupta AS. Causes and prevalence of factors causing infertility in a public health facility. J Hum Reprod Sci. 2019;12(4):287–293. doi:10.4103/jhrs.JHRS_140_18
4. Cooney LG, Dokras A. Beyond fertility: polycystic ovary syndrome and long-term health. Fertil Steril. 2018;110(5):794–809. doi:10.1016/j.fertnstert.2018.08.021
5. Dumesic DA, Meldrum DR, Katz-Jaffe MG, Krisher RL, Schoolcraft WB. Oocyte environment: follicular fluid and cumulus cells are critical for oocyte health. Fertil Steril. 2015;103(2):303–316. doi:10.1016/j.fertnstert.2014.11.015
6. Chang HM, Qiao J, Leung PC. Oocyte-somatic cell interactions in the human ovary-novel role of bone morphogenetic proteins and growth differentiation factors. Hum Reprod Update. 2016;23(1):1–18. doi:10.1093/humupd/dmw039
7. Li Y, Li RQ, Ou SB, et al. Increased GDF9 and BMP15 mRNA levels in cumulus granulosa cells correlate with oocyte maturation, fertilization, and embryo quality in humans. Reprod Biol Endocrinol. 2014;12:81. doi:10.1186/1477-7827-12-81
8. Pogrmic-Majkic K, Samardzija Nenadov D, Stanic B, et al. T-2 toxin downregulates LHCGR expression, steroidogenesis, and cAMP level in human cumulus granulosa cells. Environ Toxicol. 2019;34(7):844–852. doi:10.1002/tox.22752
9. Sreerangaraja Urs DB, Wu WH, Komrskova K. Mitochondrial function in modulating human granulosa cell steroidogenesis and female fertility. Int J Mol Sci. 2020;21(10):3592. doi:10.3390/ijms21103592
10. Wang T, Liu Y, Lv M, et al. miR-323-3p regulates the steroidogenesis and cell apoptosis in polycystic ovary syndrome (PCOS) by targeting IGF-1. Gene. 2019;683:87–100. doi:10.1016/j.gene.2018.10.006
11. Zhang CL, Wang H, Yan CY, Gao XF, Ling XJ. Deregulation of RUNX2 by miR-320a deficiency impairs steroidogenesis in cumulus granulosa cells from polycystic ovary syndrome (PCOS) patients. Biochem Biophys Res Commun. 2017;482(4):1469–1476. doi:10.1016/j.bbrc.2016.12.059
12. Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–691. doi:10.1038/s41576-019-0158-7
13. Chen LL. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat Rev Mol Cell Biol. 2020;21(8):475–490. doi:10.1038/s41580-020-0243-y
14. Huang A, Zheng H, Wu Z, Chen M, Huang Y. Circular RNA-protein interactions: functions, mechanisms, and identification. Theranostics. 2020;10(8):3503–3517. doi:10.7150/thno.42174
15. Ma Z, Zhao H, Zhang Y, Liu X, Hao C. Novel circular RNA expression in the cumulus cells of patients with polycystic ovary syndrome. Arch Gynecol Obstet. 2019;299(6):1715–1725. doi:10.1007/s00404-019-05122-y
16. Che Q, Liu M, Xu J, et al. Characterization of circular RNA expression profiles in cumulus cells from patients with polycystic ovary syndrome. Fertil Steril. 2019;111(6):1243–1251.e1241. doi:10.1016/j.fertnstert.2019.02.023
17. Zhang C, Liu J, Lai M, et al. Circular RNA expression profiling of granulosa cells in women of reproductive age with polycystic ovary syndrome. Arch Gynecol Obstet. 2019;300(2):431–440. doi:10.1007/s00404-019-05129-5
18. Liu J, Ding J. CircPSMC3 alleviates the symptoms of PCOS by sponging miR-296-3p and regulating PTEN expression. J Cell Mol Med. 2020;24(18):11001–11011. doi:10.1111/jcmm.15747
19. Lu J, Xue Y, Wang Y, et al. CiRS-126 inhibits proliferation of ovarian granulosa cells through targeting the miR-21-PDCD4-ROS axis in a polycystic ovarian syndrome model. Cell Tissue Res. 2020;381(1):189–201. doi:10.1007/s00441-020-03187-9
20. Wu G, Xia J, Yang Z, Chen Y, Jiang W, Yin T. CircASPH promotes KGN cells proliferation through miR-375/MAP2K6 axis in Polycystic Ovary Syndrome. J Cell Mol Med. 2020. doi:10.1111/jcmm.16231
21. Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinform. 2008;9(1):559. doi:10.1186/1471-2105-9-559
22. Huang X, Liu C, Hao C, et al. Identification of altered microRNAs and mRNAs in the cumulus cells of PCOS patients: miRNA-509-3p promotes oestradiol secretion by targeting MAP3K8. Reproduction. 2016;151(6):643–655. doi:10.1530/REP-16-0071
23. Li J, Chen H, Gou M, et al. Molecular features of polycystic ovary syndrome revealed by transcriptome analysis of oocytes and cumulus cells. Front Cell Dev Biol. 2021;9:735684. doi:10.3389/fcell.2021.735684
24. Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi:10.1093/nar/gkv007
25. Xu F, Liu R, Cao X. Hyperandrogenism stimulates inflammation and promote apoptosis of cumulus cells. Cell Mol Biol. 2017;63(10):64–68. doi:10.14715/cmb/2017.63.10.10
26. Li XN, Wang ZJ, Ye CX, Zhao BC, Li ZL, Yang Y. RNA sequencing reveals the expression profiles of circRNA and indicates that circDDX17 acts as a tumor suppressor in colorectal cancer. J Exp Clin Cancer Res. 2018;37(1):325. doi:10.1186/s13046-018-1006-x
27. Peng HH, Wen YG. CircDDX17 acts as a competing endogenous RNA for miR-605 in breast cancer progression. Eur Rev Med Pharmacol Sci. 2020;24(12):6794–6801. doi:10.26355/eurrev_202006_21668
28. Ren TJ, Liu C, Hou JF, Shan FX. CircDDX17 reduces 5-fluorouracil resistance and hinders tumorigenesis in colorectal cancer by regulating miR-31-5p/KANK1 axis. Eur Rev Med Pharmacol Sci. 2020;24(4):1743–1754. doi:10.26355/eurrev_202002_20351
29. Zhang W, Xu C. Circ-ELF2 Acts as a Competing Endogenous RNA to Facilitate Glioma Cell Proliferation and Aggressiveness by Targeting MiR-510-5p/MUC15 Signaling. Onco Targets Ther. 2020;13:10087–10096. doi:10.2147/OTT.S275218
30. He K, Qu H, Wang H, Zhang S, Qian XH, Regulated LW. Functional Expression of the Corepressor MTA3 in Rodent Testis. Endocrinology. 2016;157(11):4400–4410. doi:10.1210/en.2016-1213
31. Gao GZ, Zhao Y, Li HX, Li W. Bisphenol A-elicited miR-146a-5p impairs murine testicular steroidogenesis through negative regulation of Mta3 signaling. Biochem Biophys Res Commun. 2018;501(2):478–485. doi:10.1016/j.bbrc.2018.05.017
32. Xu ZF, Sun XK, Chen G, Han C, Wang F, Zhang YD. Oroxyloside inhibits human glioma progression by suppressing proliferation, metastasis and inducing apoptosis related pathways. Biomed Pharmacother. 2018;97:1564–1574. doi:10.1016/j.biopha.2017.09.100
33. Bolger-Munro M, Choi K, Scurll JM, et al. Arp2/3 complex-driven spatial patterning of the BCR enhances immune synapse formation, BCR signaling and B cell activation. Elife. 2019;8:e44574. doi:10.7554/eLife.44574
34. Jing X, Peng J, Dou Y, et al. Macrophage ERα promoted invasion of endometrial cancer cell by mTOR/KIF5B-mediated epithelial to mesenchymal transition. Immunol Cell Biol. 2019;97(6):563–576. doi:10.1111/imcb.12245
35. Chaudhuri E, Dash S, Balasubramaniam M, Padron A. The HIV-1 capsid-binding host factor CPSF6 is post-transcriptionally regulated by the cellular microRNA miR-125b. J Biol Chem. 2020;295(15):5081–5094. doi:10.1074/jbc.RA119.010534
36. Lin Q, Cai J, Wang QQ. The Significance of Circular RNA DDX17 in Prostate Cancer. Biomed Res Int. 2020;2020:1878431. doi:10.1155/2020/1878431
37. Long H, Sun B, Cheng L, et al. miR-139-5p Represses BMSC Osteogenesis via Targeting Wnt/β-Catenin Signaling Pathway. DNA Cell Biol. 2017;36(8):715–724. doi:10.1089/dna.2017.3657
38. Zhu Y, Zhao H, Feng L, Xu S. MicroRNA-217 inhibits cell proliferation and invasion by targeting Runx2 in human glioma. Am J Transl Res. 2016;8(3):1482–1491.
39. Huang W, Lu Y, Wang F, Huang X, Yu Z. Downregulation of circular RNA hsa_circ_0000144 inhibits bladder cancer progression via stimulating miR-217 and suppressing RUNX2 expression. Gene. 2018;678:337–342. doi:10.1016/j.gene.2018.08.036
40. Papamentzelopoulou M, Mavrogianni D, Dinopoulou V, et al. Detection of RUNX2 gene expression in cumulus cells in women undergoing controlled ovarian stimulation. Reprod Biol Endocrinol. 2012;10:99. doi:10.1186/1477-7827-10-99
41. Zhao F, Wang N, Yi Y, et al. Knockdown of CREB3/Luman by shRNA in Mouse Granulosa Cells Results in Decreased Estradiol and Progesterone Synthesis and Promotes Cell Proliferation. PLoS One. 2016;11(12):e0168246. doi:10.1371/journal.pone.0168246
42. Qiao GY, Dong BW, Zhu CJ, Yan CY, Chen BL. Deregulation of WNT2/FZD3/β-catenin pathway compromises the estrogen synthesis in cumulus cells from patients with polycystic ovary syndrome. Biochem Biophys Res Commun. 2017;493(1):847–854. doi:10.1016/j.bbrc.2017.07.057
43. Zhang X, Xiao H, Zhang X, et al. Decreased microRNA-125b-5p disrupts follicle steroidogenesis through targeting PAK3/ERK1/2 signalling in mouse preantral follicles. Metabolism. 2020;107:154241. doi:10.1016/j.metabol.2020.154241
44. Xu R, Qin N, Xu X, Sun X, Chen X, Zhao J. Inhibitory effect of SLIT2 on granulosa cell proliferation mediated by the CDC42-PAKs-ERK1/2 MAPK pathway in the prehierarchical follicles of the chicken ovary. Sci Rep. 2018;8(1):9168. doi:10.1038/s41598-018-27601-z
45. Savastano S, Orio F
46. Kaur S, Archer KJ, Devi MG, Kriplani A, Strauss JF
47. Takaesu G, Kishida S, Hiyama A, et al. TAB2, a novel adaptor protein, mediates activation of TAK1 MAPKKK by linking TAK1 to TRAF6 in the IL-1 signal transduction pathway. Mol Cell. 2000;5(4):649–658. doi:10.1016/S1097-2765(00)80244-0
48. Uma J, Muraly P, Verma-Kumar S, Medhamurthy R. Determination of onset of apoptosis in granulosa cells of the preovulatory follicles in the bonnet monkey (Macaca radiata): correlation with mitogen-activated protein kinase activities. Biol Reprod. 2003;69(4):1379–1387. doi:10.1095/biolreprod.103.017897
49. Fox CW, Zhang L, Sohni A, et al. Inflammatory stimuli trigger increased androgen production and shifts in gene expression in theca-interstitial cells. Endocrinology. 2019;160(12):2946–2958. doi:10.1210/en.2019-00588
50. Kim JH, Cox ME, Wasan KM. Effect of simvastatin on castration-resistant prostate cancer cells. Lipids Health Dis. 2014;13:56. doi:10.1186/1476-511X-13-56
51. Rashidi B, Abediasl J, Tehraninejad E, Rahmanpour H, Sills ES. Simvastatin effects on androgens, inflammatory mediators, and endogenous pituitary gonadotropins among patients with PCOS undergoing IVF: results from a prospective, randomized, placebo-controlled clinical trial. J Investig Med. 2011;59(6):912–916.
52. Elemam NM, Hasswan H, Aljaibeji H. Circulating Soluble ACE2 and Upstream microRNA Expressions in Serum of Type 2 Diabetes Mellitus Patients. Int J Mol Sci. 2021;22(10):5263.
53. Rubie C, Zimmer J, Lammert F, et al. MicroRNA-496 and Mechanistic Target of Rapamycin Expression are Associated with Type 2 Diabetes Mellitus and Obesity in Elderly People. Ann Nutr Metab. 2019;74(4):279–286. doi:10.1159/000499576
54. Zhang Y, Xiao H-Q, Wang Y, Yang Z-S, Dai L-J, Xu Y-C. Xu YC. Differential expression and therapeutic efficacy of microRNA-346 in diabetic nephropathy mice. Exp Ther Med. 2015;10(1):106–112. doi:10.3892/etm.2015.2468
55. Wei H, Huang L, Wei F, et al. Up-regulation of miR-139-5p protects diabetic mice from liver tissue damage and oxidative stress through inhibiting Notch signaling pathway. Acta Biochim Biophys Sin. 2020;52(4):390–400. doi:10.1093/abbs/gmaa008
56. Li J, Liu B, Xue H, Zhou QQ, Peng L. miR-217 is a useful diagnostic biomarker and regulates human podocyte cells apoptosis via targeting TNFSF11 in membranous nephropathy. Biomed Res Int. 2017;2017:2168767. doi:10.1155/2017/2168767
57. Che Q, Liu M, Zhang D, et al. Long Noncoding RNA HUPCOS Promotes Follicular Fluid Androgen Excess in PCOS Patients via Aromatase Inhibition. J Clin Endocrinol Metab. 2020;105(4):dgaa060. doi:10.1210/clinem/dgaa060
58. Lin J, Xia J, Zhang T, Zhang K, Yang Q. Genome-wide profiling of microRNAs reveals novel insights into the interactions between H9N2 avian influenza virus and avian dendritic cells. Oncogene. 2018;37(33):4562–4580. doi:10.1038/s41388-018-0279-z
59. Zhang T, Tian F, Huo R, Tang A, Zeng Y. Detection of dendritic cells and related cytokines in follicular fluid of patients with polycystic ovary syndrome. Am J Reprod Immunol. 2017;78(3):e12717. doi:10.1111/aji.12717
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.