Back to Journals » OncoTargets and Therapy » Volume 13
Vital and Distinct Roles of H2A.Z Isoforms in Hepatocellular Carcinoma
Authors Tang S, Huang X, Wang X, Zhou X, Huang H, Qin L, Tao H, Wang Q, Tao Y
Received 26 December 2019
Accepted for publication 9 April 2020
Published 18 May 2020 Volume 2020:13 Pages 4319—4337
DOI https://doi.org/10.2147/OTT.S243823
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sanjay Singh
Shaomei Tang,1,* Xiaoliang Huang,2,* Xi Wang,3,* Xianguo Zhou,4 Huan Huang,3 Liwen Qin,3 Hongyu Tao,3 Qiuyan Wang,3– 5 Yuting Tao3,6
1The First Affiliated Hospital of Guangxi Medical University, Nanning, Guangxi, People’s Republic of China; 2Department of Gastrointestinal Surgery, Guangxi Medical University Cancer Hospital, Nanning, Guangxi 530021, People’s Republic of China; 3Center for Genomic and Personalized Medicine, Guangxi Medical University, Nanning, Guangxi, People’s Republic of China; 4Department of Research, Guangxi Medical University Cancer Hospital, Nanning, Guangxi 530021, People’s Republic of China; 5Guangxi Colleges and Universities Key Laboratory of Biological Molecular Medicine Research, Nanning, Guangxi, People’s Republic of China; 6Guangxi Key Laboratory for Genomic and Personalized Medicine, Guangxi, Collaborative Innovation Center for Genomic and Personalized Medicine, Nanning, Guangxi, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Yuting Tao; Qiuyan Wang
Center for Genomic and Personalized Medicine, Guangxi Medical University, Nanning, Guangxi, People’s Republic of China
Email [email protected]; [email protected]
Purpose: H2A.Z is an oncogenic histone variant that is overexpressed in cancers. Two isoforms of H2A.Z, H2AFZ and H2AFV, are identical except for a three-amino acid difference. However, their isoform-specific functions remain unclear in cancer development. Thereby, this study aimed to investigate whether the two isoforms play distinct functions in hepatocarcinogenesis.
Materials and Methods: Expressions of H2A.Z isoforms in 116 paired hepatocellular cancerous and para-cancerous tissues were detected by employing qPCR. GEO and TCGA databases were used to probe expressions and prognostic value of the two H2A.Z isoforms. A comprehensive meta-analysis was conducted. Furthermore, co-expressed analysis of H2AFZ and H2AFV was performed by using cBioPortal database. H2A.Z binding genes from Chip-seq were intersected with H2A.Z isoforms co-expressed genes to perform functional annotations. Cell proliferation experiments from H2AFZ knockout HepG2 and BEL-7402 cells were implemented. Finally, RNA-seq was applied to analyse alternative splicing in H2AFZ knockout and wild-type cells.
Results: H2AFZ and H2AFV were both significantly upregulated (P < 0.01) in hepatocellular carcinoma and related to poor prognosis (P < 0.01). The two H2A.Z isoforms played vital roles in cell proliferation. It is also predicted that unique functions of H2AFV contain spindle midzone and microtubule, while H2AFZ is especially associated with RNA export and spliceosome. Further, devoid H2AFZ may restrain liver cancer cell proliferation and cause many alternative splicing events.
Conclusion: Both H2A.Z isoforms play vital and distinct roles in the occurrence and progression of liver cancer, which may pave a way for novel therapeutic applications for cancers in the future.
Keywords: H2A.Z, H2AFV, H2AFZ, hepatocellular carcinoma, functional annotations, distinct roles
Introduction
Cancer is believed to be a leading cause of mortality in the 21st century and a main obstacle in improving life expectancy worldwide. It is estimated that liver cancer is the sixth most common cancer and the fourth leading cause of death in the world in 2018, with an annual increase of 841,000 cases and 782,000 deaths. Among primary liver cancers, hepatocellular carcinoma (HCC) is the most frequent (75%–85%).1 More than half of the new cases and deaths due to liver cancers occur in China, with an increase of 466,100 cases and 422,100 deaths in 2015. To make matters worse, HCC brings about a tremendous economic burden and exhibits poor prognosis owing to the failure of early diagnosis and effective treatment.2 Hence, it is crucial to gain an insight into the role of the pathogenesis of HCC and develop novel prognostic predictors and targeted agents.
Recent research on the mechanisms of HCC has elucidated the importance of epigenetic regulation.3 Multiple omics technologies and large-scale parallel sequencing have been employed to shed light on epigenetic events, such as histone methyltransferase, SETDB1,4 histone variant macroH2A,5 EZH26 and BRD4.7 Among these, histone variants and their chaperones are emerging as key regulators in cancers.
H2A.Z, a highly conserved variant of H2A, with 60% similar to canonical histone H2A in eukaryotic cells.8 H2A.Z has two distinct isoforms, H2A.Z.1 and H2A.Z.2 (called as H2AFZ and H2AFV, respectively), which are encoded by two non-allelic genes and driven by independent promoters on distinct chromosomes, albeit they only differ in three amino acids at the protein level.9,10 These three amino acids are far apart on these polypeptides; however, the two proteins are structurally similar to each other. Hence, there are no antibodies to distinguish between H2AFZ and H2AFV,11 and their isoform-specific functions remain poorly defined. Studies on H2AFZ knockout mouse revealed that H2AFZ and H2AFV are distinct and non-redundant.12 H2A.Z has been observed to be overexpressed in breast cancer,13 prostate cancer,14 bladder cancer,15 and lung cancer.16 Nevertheless, these studies laid emphasis only on H2AFZ or did not clearly discriminate between the two isoforms.
It should be noted that in metastatic melanoma,17 both H2AFZ and H2AFV share genomic occupancy patterns and interact with similar histone chaperones, whereas only H2AFV loss can result in dramatic changes of chromatin structure by impairing BRD2 and E2F1 functions and affect melanoma drug sensitivity, which clearly act differently from H2AFZ. By contrast, Yang et al show that H2AFZ is significantly upregulated and pertinent with poor prognosis in HCC patients; yet H2AFV exhibits no changes between tumor and non-tumor patients. They consider that H2AFZ modulates cell cycle and epithelial–mesenchymal transition (EMT) regulatory proteins to target H2A.Z decomposition in HCC.18
Unexpectedly, in the present study, we demonstrated that not only H2AFZ but also H2AFV was significantly upregulated in HCC compared to normal tissue. We also found that patients exhibiting high expression of H2AFZ or H2AFV were both significantly associated with shorter overall survival and disease-free survival. More importantly, our results indicated that both H2A.Z isoforms played vital roles in cell proliferation; however, a fraction of functions between them were distinctive. Specifically, H2AFZ associated genes were enriched in RNA splicing and exporting, while H2AFV associated genes focused on microtubule depolymerization, spindle midzone and chromatin binding. Collectively, this study suggests that the two H2A.Z isoforms both have oncogenic potential in hepatocarcinogenesis; nevertheless, they may play distinct and non-redundant roles in HCC, which may pave a way for researches related to the functional and evolutionary specialization of these isoforms in the future.
Materials and Methods
Patient Samples
One hundred and sixteen paid tissue samples were obtained from patients at Guangxi Medical University Cancer Hospital in China from December 2017 to August 2019. Two experienced pathologists authenticated histopathological results. And clinical data included age, gender, Barcelona Clinical Liver Cancer System (BCLC) and whether RNA-seq or not (Supplemental Table 1). All tissues are preserved by the central laboratory in accordance with standard procedures. The study was approved by the Ethics and Human Subject Committee of Guangxi Medical University. All experiments and methods were performed according to relevant guidelines and regulations.
Data Collection
A search of H2AFZ and H2AFV in liver cancer was conducted in the GEO database (http://www.ncbi.nlm.nih.gov/geo/) with the following keywords: (hepatocellular OR liver OR hepatic OR hepatoma OR HCC OR hepatoma OR Hepatoblastoma) AND (malignan*’ or “cancer” or “tumor” or “tumour” or “neoplas*” or “carcinoma”). The entry type was restricted to “series,” the study type was set as “Expression profiling by array” and “Expression profiling by high throughput sequencing”, and the organism was filtered by “Homo sapiens.” The criteria for inclusion were as follows: (1) pathological diagnosis of patients with liver cancer; (2) cancerous and noncancerous samples were involved; (3) noncancerous and cancerous samples included at least two samples in the form of tissue; (4) expression profiling data for H2AFZ or H2AFV were available; (5) and the preoperative therapy (including radiotherapy, chemotherapy, or chemoradiotherapy) was not performed. For eligible study, expression profiles and annotation files of samples were downloaded. Moreover, RNA sequencing (RNA-seq) expression profiles of HCC and noncancerous samples and corresponding clinical information were downloaded from the Genomic Data Commons data portal (https://portal.gdc.cancer.gov/).
Tissue Digestion
Complete media was prepared with 90%FBS (BI Biological) and 10% DMSO. The tumor tissues were each minced with scissors in 6-cm plates and digested in 2.5mg/mL collagenase type Ⅱ (Life Technologies) for 40 minutes at 37°C.The cells were filtered through a 70 μm cell strainer, lysed in RBC buffer (Solarbio), washed, resuspended in complete media and stored at −80°C.
RNA Isolation and Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA was distilled from frozen tissues with a Trizol Reagent (Invitrogen) and then synthesized cDNA using High-Capacity cDNA Reverse Transcription Kits (Thermo Fisher Scientific) according to the manufacturer’s instructions. Quantifications of H2AFV, H2AFZ and Actin were conducted using the TB GreenTM Premix Ex TaqTM II Kit (Takara) in the StepOnePlus real-time PCR system (Applied Biosystems) based on the manufacturer’s instructions. Actin was set as the internal reference. The sequences for the Actin, H2AFV and H2AFZ primers are as follows: H2AFZ 5′- GCA GTT TGA ATC GCG GTG −3′ (forward) and 5′- GAG TCC TTT CCA GCC TTA CC −3′ (reverse); H2AFV 5′- GGA GTC AGA TTC AAA GGA −3′ (forward) and 5′- TCA AGG CAT CAG GTA AGG −3′ (reverse); Actin F-CTC CAT CCT GGC CTC GCT GT and R-GCT GTC ACC TTC ACC GTT CC;Relative expression values of H2AFV and H2AFZ were calculated by using formula control −2-(Target Ct−Control Ct).
Bioinformatics
Co-expressed genes of H2AFV or H2AFZ were obtained from prediction websites cBioPortal (Cerami et al 2012; Gao et al 2013) (http://www.cbioportal.org/). KEGG Orthology-Based Annotation System (KOBAS) (http://kobas.cbi.pku.edu.cn/) and The Database for Annotation, Visualization and Integrated Discovery (DAVID) (https://david.ncifcrf.gov/) were, respectively, used to conduct Pathways and GO annotations including Biological Process (BP), Cellular Component (CC) and Molecular Function (MF). GSEA 3.0 software was used to perform Gene Set Enrichment Analysis.19 HTSeq-FPKM mRNA expression data of 108 patients were imported into the GSEA software. Parameters were set as follows: gene sets database: c2.cp.kegg.v7.0.symbols.gmt; number of permutations: 1000; collapse dataset to gene symbols: False; enrichment statistic: weighted, metric for ranking genes: Signal2Noise. Based on the average expression of H2AFZ, these samples were divided into high expression groups (n=46) and low expression groups (n=62). It is statically significant when P values less than 0.05. Intersected genes were analysed by a website BioVenn (http://www.biovenn.nl/index.php).
Cell Culture
HepG2 cells were purchased from Cell bank of Chinese Academy of Sciences, and BEL-7402 cells were purchased from Shanghai Zhong Qiao Xin Zhou Biotechnology Company. The two cell lines were purchased commercially and were cultivated according to the protocols from their supplier. All cell lines were grown in DMEM complete medium (Gibco by Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (Biological Industries Israel Beit Haemek), and were cultured in an incubator of 37°C and 5% CO2.
Establishment of H2AFZ Knockout Sublines from Human Liver Cancer Cell Lines
The primers targeted for CRISPR-Cas9 genome modification were designed employing the online design tool (http://crispr.mit.edu/). The following are the guide RNA sequences for H2AFZ knockout: H2AFZ(I1)-F CACCGAGACGCTCGATGACTCCGC; H2AFZ(I1)-R AAACGCGGAGTCATCGAGCGTCTC; H2AFZ(I3)-F CACCGAGGCCTAAGAGAACGCTA; H2A.Z(I3)-R AAACCTAGCGTTCTCTTAGGCCTC. The annealed sgRNAs were ligated into the BbsI-digested pSpCas9(BB)-2A-Puro (pX459) V2.0 plasmid (Adgene, Cambridge, MA). Insert sequence is finally validated. In line with instructions, HepG2 and BEL-7402 cells (1.8 × 106) were reverse transfected with 1 ug each of gRNA constructs applying Lipofectamine LTX® reagent (Thermo Fisher Scientific). Selected individual colonies were amplified, screened and verified by Western blotting.
Western Blotting
Cells were lysed using RIPA lysis Buffer, collected and stored at −80°C. Protein concentration was determined by the Bradford assay (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Equal amounts (30μg) of proteins were separated by 14% SDS-PAGE and transferred onto polyvinylidene difluoride membranes for 1.5 h. The membranes were washed one time with TBS/0.1% Tween-20 (TBST) buffer and incubated with a solution containing the primary antibody (1:1, 000) at 4°C overnight. Then, the membranes were washed three times with TBST, and incubated with a solution containing the horseradish peroxidase (HRP)-conjugated secondary antibody (1:3000) for 1.5h at 20–25°C.Following incubation, the membranes were washed three times with TBST. Enhanced chemiluminescence (ECL) was used to detect the immunoreactive bands, according to the manufacturer’s recommendation.
Experiments and Bioinformatic Analysis of CHROMATIN Immunoprecipitation Coupled with High-Throughput Sequencing (Chip-seq)
HepG2 and BEL-7402 cells were prepared for ChIP-seq as previously described with modifications. Briefly, protein-DNA complexes were cross-linked with 1% formaldehyde for 10 minutes, followed by quenching with 0.125M glycine for 5 minutes. Sonication was performed using Bioruptor Plus with 10 pulses consisting of a 30-second sonication followed by a 30-second rest at 25% amplitude on tube coolers to yield a range of products between 100–600p. Sonicated chromatin was collected for 15 min at 4°C. Immunoprecipitation used Anti-Histone H2A.Z antibody [EPR6171(2)(B)] - ChIP Grade ab150402. Cross-links were reversed overnight at 65°C, followed by purification of the enriched DNA using manual operation.
For H2A.Z Chip-seq, two biological replicates of HepG2 or 7402 cells were performed. Library preparation was performed according to the published protocol, including DNA-end repair, PCR amplification and qualified library for sequencing. Paired-end sequencing was constructed on Illumina’s NextSeq 500 platform. For sequence analysis, quality control was conducted with FastQC, and reads were aligned to the human genome (hg19) using Bowtie2. Masc2 was used to peak calling, and Chipseeker was conducted for annotations and visualization of peaks.
Experiments and Bioinformation Analysis of RNA-Seq
Knockout of H2AFZ and Wild-type in HepG2 or BEL-7402 cells were performed RNA-seq. Also, 108 hepatocellular carcinoma tissues from 116 samples were carried out RNA-seq. Total RNA was extracted from the HepG2 and BEL-7402 cells. Then, the integrity of the total RNA was determined by 2100 Bioanalyser (Agilent) and quantified using the NanoDrop (Thermo Scientific). RNA purification, reverse transcription, library construction and sequencing (2x150 paired-end) were performed at WuXi NextCODE in Shanghai according to the manufacturer’s instructions (Illumina).
For bioinformation analysis, quality control was conducted with FastQC, and reads were aligned to the human genome (hg19) using Salmon. rMATS 4.0 was used to obtain alternative splicing events. And R statistical software (version 5.3) was drawn UpSet plot.
Cell Growth and Proliferation Assay
MTT experiments were used to assess cell viability. Cells (1×103 cells/well) were seeded in 96-well plates, and cultured in an incubator of 37°C and 5% CO2 for 1 to 8 days. The supernatant in each well was then replaced with 90 μL medium and 10 μL MTT solution, and the cells were incubated at 37°C for 4h. The absorbance was detected at 450nm. Cells (400 cells/well) were seeded into 6-well plates and cultured in an incubator of 37°C and 5% CO2 for 14 days. Then, the cells were washed with PBS, fixed 100% methanol for 30 minutes at room temperature, and stained with 0.1% crystal violet for 30 minutes at room temperature. Following that, cells were washed with PBS three times and counted. Flow cytometry was performed to detect the cell cycle. The wild-type group and H2A.Z knockout group cells (2×105) were counted and seeded in 6-well plates, then cultured with Tdy for 18h for the first time and cultured with Tdy for 16h for the second time. The cells were harvested and fixed in 70% ethanol at 4°C overnight. Washing fixed cells with PBS for one time. Then, add 500μL PI and incubate for 30 minutes at room temperature in the dark. After incubating, these cells were analysed by flow cytometer (BD Biosciences, San Jose, CA, USA).
Statistical Analyses and Comprehensive Meta-Analysis
Student’s T-tests were used to compare the difference between groups. Error bars denote ±standard deviation except where noted. The relationship between H2A.Z expression level and clinicopathological features was assessed by using Chi-square test. The survival significance of H2AFV and H2AFZ in HCC was elucidated using Kaplan–Meier plot and Log-rank test in a TCGA cohort. The receiver operating characteristic (ROC) curve was conducted to pinpoint the diagnostic efficiency of H2AFV and H2AFZ in HCC. The optimum cutoff point was set with the Youden index (sensitivity + specificity − 1). All results were constructed using SPSS 22.0 and graphs were gained through GraphPad Prism 5.0. P value less than 0.05 was indicated as a statistically significant difference.
After the expression values, H2AFZ and H2AFV were log2-transformed, the mean (M) and standard deviation (SD) for each cancerous and noncancerous group were calculated. In addition, Stata 12.0 software was used for performing a meta-analysis of data aggregated from different datasets. When the pooled SMD >0, the gene expression in HCC was higher than that in the noncancerous group; when SMD was <0, the gene expression was higher in the noncancerous group than that in HCC. The expression of the H2AFZ and H2AFV in the cancerous and noncancerous samples was displayed on forest plots. The chi-squared test of Q and the I2 statistic were calculated to assess heterogeneity across the studies and to determine the appropriateness of applying either a random effects model or fixed effects model to the pooling process.
Results
H2AFZ and H2AFV Were Both Aberrantly Upregulated in HCC Patients and Associated with Advanced Stage and Poor Prognosis
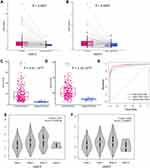
To measure expressions of H2A.Z, we detected mRNA expression of H2A.Z isoforms in 116 pairs of cancer and adjacent samples. We found that not only H2AFZ expression was predominantly increased in the 116 HCC tissue samples than theses non-cancerous liver tissue samples (P < 0.0001) (Figure 1A), but also, very interestingly, expression of H2AFV in HCC was significantly ascended (P = 0.0007, Figure 1B).
The previous study reported that only H2AFZ, but not H2AFV, was significantly overexpressed in HCC based on four different HCC datasets from Gene Expression Omnibus (GEO) database.18 However, four datasets only occupy a small part of liver cancer-associated datasets in GEO database and the criteria for inclusion have not been reported, which could lead to selection bias. Thereby, we analyzed the expression of H2AFZ and H2AFV in HCC based on TCGA data. As shown in Figure 1C and D, expressions of the two H2A.Z isoforms were significantly increased in HCC. Besides, we demonstrated that both H2A.Z isoforms had statistical significance in stages (Figure 1E and F). We also analyzed the sensitivity and specificity of H2AFZ and H2AFV based on TCGA data by receiver operator characteristic (ROC) curve to assess their diagnostic value. The area under the curve (AUC) of H2AFZ was 0.96 and H2AFV was 0.93. A greater AUC was acquired after the combination of H2AFZ and H2AFV (Figure 1G). To comprehensively evaluate H2A.Z isoforms in liver cancer, a meta-analysis was performed. Including in-house PCR data, a total of 62 datasets were included in the present study. The features of the included datasets are depicted in Supplement Table 2. Among the 62 datasets, GSE10143 only contained the expression of H2AFZ (Supplemental Table 2). Thus, there were 62 datasets contained the expression of H2AFZ and 61 datasets contained the expression of H2AFV. In total, 3016 HCC samples and 2281 noncancerous samples were included for the integrated meta-analysis. We first performed a meta-analysis to evaluate the expression of H2AFZ in liver cancer based on the 62 datasets. Given the apparent heterogeneity (p < 0.05, I2 = 82.3%), a random effects model was applied, and remarkable up-regulation (SMD = 1.22; 95% CI: 1.05–1.40; P<0.001) of H2AFZ was found in the HCC groups (Figure 2). We also performed a meta-analysis to evaluate the expression of H2AFV. Significant heterogeneity was observed (p < 0.05, I2 = 81.3%) and random effects model was applied to merge the results. As shown in Figure 3, H2AFV was also significantly overexpressed in HCC tissues (SMD = 0.76; 95% CI: 0.59–0.93; P<0.001). In short, both H2AFZ and H2AFV had a meaningful overexpression in HCC.
 |
Figure 2 Forest plot of 62 data sets and in-house PCR data for the H2AFZ expression in liver cancer (with random-effect model). |
 |
Figure 3 Forest plot of 61 data sets and in-house PCR data for the H2AFV expression in liver cancer (with random-effect model). |
Next, we analyzed the association between expression of H2A.Z isoforms and clinical parameters to explore the clinical significance of H2A.Z in HCC based on TCGA data. As shown in Table 1, H2AFZ and H2AFV were both significantly associated with serous alpha-fetoprotein levels, tissue grading, pathological staging, and T category. We further analyzed the association between the expression of each H2A.Z isoform and prognosis. Both H2A.Z isoforms had a similar tendency that high expression was significantly associated with shorter overall survival and disease-free survival (Figure 4). In brief, these results indicated that both H2AFZ and H2AFV play an important role in tumorigenesis and progression.
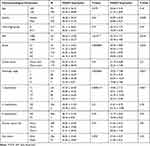 |
Table 1 The Relationship Between H2AFZ, H2AFV and HCC Clinical Features |
Both H2A.Z Isoforms Had Similar and Distinct Functions in HCC
In this study, the two H2A.Z isoforms were significantly overexpressed in HCC and correlated to diagnosis and prognosis, which implied their important roles in hepatocarcinogenesis. Interestingly, certain of researches uncovered that functions of the two H2A.Z isoforms are not-redundant and distinct.10,20-22 Thereby, it is fascinating to explore whether they give scope to identical functions in HCC occurrence and development. We predicated co-expressed genes of H2AFZ and H2AFV by employing cBioPortal database. A meaningful number, 2372, of genes with expression patterns that prominently cognate with H2AFZ and 569 genes with H2AFV (P < 0.001, r > 0.3 or r < −0.3), were further investigated the latent molecular mechanisms of similarities and difference. Functional analysis revealed that all of the significant pathways from 569 H2AFV co-expressed genes were overlapped pathways from 2372 H2AFZ co-expressed genes (Corrected P-value <0.05) (Figure 5A), and most of GO terms including Biological Process (BP), Cellular Component (CC) and Molecular Function (MF) were identical (Supplemental table 3), for instance, cell cycle and division. Yet a handful of GO annotations were extraordinary, as shown in Supplemental tables 4 and 5 (FDR <0.05). For example, protein localization to kinetochore, spindle midzone and microtubule binding were unique GO of H2AFV co-expressed genes; while mRNA splicing via spliceosome, catalytic step 2 spliceosome and RNA export were particular GO of H2AFZ co-expressed genes (Figure 5B and C). In a word, the two H2A.Z isoforms play similar roles in cell proliferation; however, to an extent, their oncogenic ways seemed likely distinctive in hepatocarcinogenesis.
For more particular knowledge of the transcriptional regulatory mechanism of H2A.Z and examining its directly binding genes, the physical association of H2A.Z with DNA was examined by H2A.Z high-throughput Chromatin Immuno-Precipitation followed by sequencing (Chip-seq) in HepG2 cells and BEL-7402 cells. A total of 4583 genes binding H2A.Z have been found in 7402 cells and 1887 genes in HepG2 cells. One thousand one hundred and seventy-eight genes were overlapped in the two cell lines (Figure 6A). Subsequently, we integrated H2A.Z binding genes with H2AFV co-expressed genes or H2AFZ genes to further decipher the roles of the two H2A.Z isoforms in hepatoma cells. For more legibly delineating these intersecting genes, we defined three classes of genes: H2A.Z binding genes from the two HCC cell lines (Class I); intersecting genes of Class I with H2AFV co-expressed genes (Class II); intersecting genes of Class I with H2AFZ co-expressed genes (Class III) (Figure 6A). Functional annotations were performed in Class II and Class III genes. The results of the KEGG pathway analysis depicted that the bulk of pathways in Class II was overlapped with Class III, except Platinum drug resistance and Pancreatic cancer (Table 2). GO annotations revealed that Class II is mainly enriched for cell cycle and division, these GO annotations also appeared in Class III (Supplemental table 6). However, special GO annotations such as microtubule depolymerization, cellular response to gamma radiation, spindle midzone and DNA binding were discovered in Class II (Figure 6B). Unlike GO annotations in Class II, Class III is particularly concentrated on mRNA export from nucleus, regulation of glucose transport and intracellular ribonucleoprotein complex (Figure 6C and Supplemental table 7). A protein–protein interaction network was constructed by using STRING database in Class II genes. A total of 5 hub genes including TOP2A, RAD51, BRCA1, ASPM and RAD54L were chose from the PPI with a degree of >10 (Figure 6D). We also performed RNA-seq in 108 hepatocellular carcinoma tissues. Results delineated that both H2AFZ and H2AFV were significantly associated with TOP2A, RAD51, BRCA1 and RAD54L (p < 0.001), except for ASPM1 (Supplemental Figure 1). Intriguingly, these four genes (TOP2A, RAD51, BRCA1 and RAD54L) were closely related to DNA double-strand break (DSB), which was repaired by two main pathways, homologous recombination and non-homologous end joining23,24 (Supplemental Figure 2). Further, based on the average expression of H2AFZ, we divided HCC samples into H2AFZ high expression groups (n=46) and low expression groups (n=62) and performed Gene Set Enrichment Analysis (GSEA). KEGG_ homologous recombination is one of the prominently enriched gene set in H2AFZ high expression groups and expressions of concerned genes in this pathway were increased in H2AFZ high expression groups (Supplemental Figure 3). Consistent with former results, the two H2A.Z isoforms associated genes were mainly enriched in cell proliferation aspects. Amazing that they also carried out distinct functions in liver cancer occurrence and process. H2AFZ might influence RNA process, and then lead to hepatocarcinogenesis, while H2AFV associated genes concentrated on microtubule and spindle midzone.
 |
Table 2 The Pathways from Intersecting Genes of Chip-Seq Genes and H2AFV Co-Expressed Genes or H2AFZ Co-Expressed Genes (Class II or Class III Genes) |
Inhibition of H2AFZ Can Control Cell Proliferation and Then Restrain Liver Cancer Occurrence and Progression
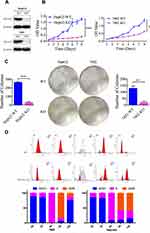
To better understand molecular functions of H2A.Z in vitro, H2AFZ was removed in HepG2 and BEL-7402 cells and the effect of knockout was confirmed by Western blot analysis (Figure 7A). MTT and colony formation assays were performed to appraise the functions of H2AFZ about growth and proliferation. Results showed that knockout of H2AFZ can inhibit cell viability and colony formation ability (P<0.05; Figure 7B and C). The anti-growth effects might be partly explicated via cell proliferation regulation, such as cell cycle. Flow cytometry analyses demonstrated that H2AFZ-knockout groups significantly increased the number of cells in G1 phase and decreased in S and G2 phases than wild-type groups (P<0.05; Figure 7D). Besides, RNA-seq with H2AFZ knockout HepG2 and BEL-7402 cells showed that part of cell cycle genes such as CDK4, CDK5, CCNQ, CDK18, CCNY, CCNE2, CDK2 is significantly attenuated in H2AFZ deficient HepG2 and BEL-7402 cells (Supplemental Figure 4). To better understand and expound the molecular mechanism of H2AFZ in hepatocarcinogenesis, we performed GSEA by RNA-seq from 108 hepatocellular carcinoma tissues. Results showed that KEGG_Cell Cycle is a highly significantly enriched gene set in H2AFZ high expression groups, and expressions of relative genes in this pathway were higher than H2AFZ low expression groups (Supplemental Figure 5). On the whole, these cell cycle genes were up-regulated in H2AFZ high expression groups. These results suggest that targeting of H2AFZ could influence the cell cycle and inhibit hepatoma cell proliferation. Consistent with functional annotations employing bioinformatics, H2AFZ can advance cell growth and proliferation in vitro, and result in liver tumorigenesis.
H2AFZ Might Affect RNA Process Employing Alternative Splicing
A limited number of protein-coding genes make it difficult to expound aplenty proteomic phenotypes.25,26 Alternative splicing (AS) is a major physiological phenomenon that can result in the coding of diversity protein via exon skipping, intron retention and so on.27,28 Previous results demonstrated that H2AFZ associated genes uniquely focused on RNA process. Hence, we would like to if H2AFZ can alter RNA splicing and lead to gene expression disorder. Results of RNA-seq showed that knockout H2AFZ in HepG2 cells could cause 1659 notable up-regulated genes and 1183 down-regulated genes. Alike, 332 genes expressions increased after knocking out H2AFZ in BEL-7402 cells and 462 genes decreased (Figure 8A and B). We took the intersection of significant differentially expressed genes in the two cell lines for functional annotations. It could be discovered that H2AFZ was associated with cell growth, proliferation, migration and histone deacetylase binding (Figure 8C and D). Furthermore, as shown in Figure 9A, after knockout H2AFZ in HepG2 cells, total 3329 splicing events in 2225 genes were significantly changed (FDR < 0.05), including 2715 Skipping Exon (SE) in 1888 genes, 273 Alternative 3ʹ splice Site (A3SS) in 243 genes, 233 Alternative 5ʹ splice Site (A5SS) in 214 genes, 289 Mutually Exclusive Exons (MXE) in 243 genes and 109 Retention Intron (RI) in 103 genes. Similar to HepG2 cells, in the absence of H2AFZ, total of 2391 splicing events in 1711 genes had a statistical significance in BEL-7402 cells. Among them, SE event was the most predominant type and RI was the rarest type (Figure 9B). These results also suggested that one gene can occur in several mRNA splicing events. Followed that, we subjected these overlapped genes (669 genes, Figure 9C) from HepG2 and BEL-7402 cells to DAVID database to retrieve the biological process. The major biological pathways were DNA repair, p53 binding, histone deacetylase binding and regulation of RNA splicing (Figure 9D). In sum, these results hinted that H2AFZ might monitor RNA process, and then affect cell growth and proliferation in liver cancer.
Discussion
Liver cancers are notoriously refractory to traditional cancer therapies; worse still, majority of the patients often emerge resistant to chemotherapy, thus leading to treatment failure.2 Hence, exploring new avenues is the need of the hour. Extensively studies have been carried out on H2A.Z in recent years, and its variants and chaperones have emerged as pivotal roles in tumorigenesis and development.13,14,17 However, many others are likely still undiscovered.29 To address these knowledge gaps, in this study, we demonstrated that the H2A.Z isoforms, H2AFZ and H2AFV, are both significantly upregulated in HCC, and associated with poor prognosis. More importantly, functional annotations from H2AFZ or H2AFV associated genes unveiled that the two H2A.Z isoforms both play vital and distinct roles in the occurrence of liver cancers and progression.
Previous studies report that H2A.Z undergoes a duplication event, resulting in two different paralogs, H2AFZ and H2AFV, which are identical except for substitutions at three amino acid residues. Although the two isoforms are similar, evidence suggests that they have distinct functions.9,30 Research presented to knockout H2AFZ or H2AFV has found that the two H2A.Z isoforms are deposited into chromatin by similar mechanisms, which suggested a redundancy to their process. Nevertheless, the two knockout lines displayed that there are differences between H2AFZ and H2AFV with regard to gene expression and acetylation. This research proves the overlapped and distinctive functions of H2A.Z isoforms.10 In this regard, a particular role of H2AFV, as a driver of malignant melanoma was reported in 2015. They demonstrated that an individual property of H2AFV is to accelerate E2F1, BRD2, and histone acetylation levels. H2AFV may act together with histone acetylation to draw co-activators and TFs, such as BRD2 and E2F1, respectively, to boost the expression of cell cycle factors, which may shed light on a latent epigenetic therapeutic dimension towards enhancing drug sensitivity.17 Subsequently, Yang et al reported that H2AFZ, but not H2AFV is highly expressed in liver cancers according to four diverse HCC datasets from the GEO database.18 However, these four datasets only occupy a minor fraction of HCC associated datasets, which could cause selection bias. Recently, a report suggests that highly expressed H2AFV, but not H2AFZ, partly rescues Floating-Harbor syndrome (FHS) phenotype due to a lack of SRCAP in FHS models, through a mechanism dependent on the amino acid residue serine 38 being substituted with a threonine in H2A.Z during nucleosomal exchange. This histone variant can change nucleosomal dynamic properties, leading to selective effects on gene expression and mediating high tissue specificity features in a developmental disorder.31 Another research reveals that H2AFZ is significantly related to schizophrenia and plays crucial roles in some biological processes, such as maintenance of heterochromatin euchromatin status and chromosome segregation.32 In brief, these studies indicate that the two isoforms of H2A.Z are non-redundant and have isoform-specific functions.
Consequently, we assessed the expression of H2A.Z isoforms in HCC from a cohort from Guangxi, China. Interestingly, both H2AFZ and H2AFV were significantly increased in HCC patients. Furthermore, based on TCGA and GEO databases, both isoforms were also significantly upregulated in HCC tissues and associated with poor prognosis. Later, co-expressed genes of H2AFZ and H2AFV, respectively, were analyzed by using cBioPortal database. Most pathways and GO annotations from the H2AFV or H2AFZ co-expressed genes were similar and mainly related to cell proliferation. However, spindle midzone and microtubule binding were unique GO terms of H2AFV co-expressed genes, while H2AFZ co-expressed genes were particularly enriched in mRNA splicing via spliceosome and RNA export. In accordance with the previous study,12 H2A.Z is associated with cell proliferation, cell cycle and DNA replication. Faast and colleagues12 discovered that H2A.Z knockout embryo of mice still proceeds division and differentiation and result in the formation of normal H2A.Z(-/-) blastocysts. It cannot observe embryonic defects until shortly after implantation. In this latter study, only H2AFZ was knocked out. This implies that H2AFV is insufficient compensation for the loss of H2AFZ in mice, especially at early stages of mouse embryo differentiation. Subsequently, Dryhurst et al cover that the two isoforms of H2A.Z display similarity genomic localization and levels of N-terminal acetylation. Yet there is a slight difference in the association of the two H2A.Z isoforms with post-translation modification. Both isoforms are widely expressed in human tissues, but they display different promoter sequences that could lead to tissue-specific and temporal differences in gene expression.9 While in this study, it is predicated that subtle differences between the two H2A.Z isoforms were related to microtubule binding and RNA export. It is reported that cells lacking H2AFV exhibited slower growth than those lacking H2AFZ, thus implying that the two subtypes of H2A.Z have different mechanisms of action on nucleosome stability and chromatin structure.10 Further, inhibition of H2A.Z, the genome is highly unstable due to defects in the chromosome segregation process, which illuminates H2A.Z is essential for the exact transmission of chromosomes.33 Another study reported that the two H2A.Z isoforms have context-specific roles that replenish each other to mediate activity-induced neuronal gene transcription.11
Clarifying where the binding sites of specific proteins across the genome is crucial for decoding gene regulatory network. Chromatin immunoprecipitation coupled with high-throughput sequencing (Chip-seq) is a widely used technique at present to determine the DNA regions binding special protein.34,35 Given that antibodies cannot differentiate between the two isoforms of H2A.Z, a series of experiments were performed on H2A.Z.29 H2A.Z-binding genes were identified from Chip-seq, which were studied for intersection with genes co-expressed in the two H2A.Z isoforms. These overlapping genes were annotated separately employing websites of KOBAS and DAVID. As shown in Figure 6B and C, specific GO annotations of H2AFZ such as regulation of glucose transport and RNA export were delineated, while microtubule depolymerization, cellular response to gamma radiation and DNA binding were uncovered only in GO enrichment of H2AFV associated genes. Besides, a protein–protein interaction network was constructed from the overlapping genes of Chip-seq and co-expressed H2AFV genes, which identified 5 core genes consisting of TOP2A, RAD51, BRCA1, ASPM, and RAD54L (Figure 6D). A research reported that RAD51 focus formation, a characteristic of recombinational repair, was dysregulated in H2AFV-devoid cells, but not in H2AFZ-devoid cells. Increasing radiation sensitivity can decrease repair activity in H2AFV-devoid cells but not in H2AFZ-devoid cells, which suggest that H2AFV is required for proper DNA repair by adjusting RAD51 focus formation.36 Moreover, H2A.Z occupancy promotes marking weak introns and inspection by the spliceosome. And in the absence of H2A.Z, overexpression of splicing ATPase Prp16 controls growth and splicing defects.37 Although H2AFZ and H2AFV have similar functions in HCC development, quite a few differences between them were hinted.
As mentioned, we further performed RNA-seq to survey the relation between H2AFZ and Alternative Splicing (AS). Consistent with conjecture, knockout of H2AFZ can lead to alterations in some AS events, and the most frequent event is Skipping Exon. Surprisingly, genes from significant AS events concentrated on functions including DNA repair, histone deacetylase binding and regulation of RNA splicing. A previous study reveals that histone reduction accelerates extensive alterations in pre-mRNA processing including retention intron and other AS events.38 A special mechanism of splicing occurs when H2A.B.3 collects RNA processing factors from the spliceosome to activate genes following the replacement of H2A.Z.39 Another evidence37 demonstrates that H2A.Z occupancy facilitates co-transcriptional splicing of suboptimal introns. In the efficient splicing of weak introns, it requires H2A.Z and Swr1 nucleosome remodeling complex. Further, H2A.Z deficiency may lead to growth and splicing defects, which is compensated by elevating splicing ATPase Prp16. Meanwhile, Neves and colleagues40 reveal ample genetic interactions between H2A.Z and U2 snRNP-associated proteins. H2A.Z boosts effective spliceosomal assembly related to the U2 snRNP, while H2A.Z quiescence causes attenuated recruitment of downstream snRNPs to infant RNA. In brief, these data illustrate H2A.Z is fundamental to spliceosome assembly and pre-mRNA splicing. Hence, we will continue to carry out researches about the association between H2A.Z and splicing in our clinical samples and experiments in the future. In spite of an important function for the H2A.Z and RNA splicing, distinct post-translation modifications (PTMs) including acetylation,41 phosphorylation,42 methylation,43 ubiquitination and SUMOylation44 are also critical for H2A.Z to modulate gene expression and occurrence of disease. For example, H2A.Z acetylation (H2A.Zac) positively associated with signal transduction.41 Also, SETD6 may mediate monomethylation of lysine 7 of H2A.Z (H2A.ZK7me1) and repress gene expression in mESCs, while dimethylation of lysine 101 (H2A.ZK101me2) is correlated to gene activation in human cells.43 In our study, knockout of H2AFZ experiments in vitro demonstrated that targeted H2A.Z inhibition can control cell growth and proliferation, which preliminary verified the importance of H2A.Z in hepatocarcinogenesis. Coincident with reported results, H2A.Z congregates around the transcription start site of cell cycle regulatory molecules in bladder cancer cells and promotes cancer-related gene expression.15
In conclusion, we demonstrated that both H2A.Z isoforms were significantly upregulated in HCC and related to poor prognosis. The two H2A.Z isoforms played essential roles in cell proliferation. Yet, unique functions of H2AFV were predicted, such as spindle midzone and microtubule, while H2AFZ may regulate RNA processing, and then affect DNA repair. Although the final mechanism remains to be determined, these findings augment our current state of knowledge regarding vital and diverse roles of H2A.Z isoforms in liver tumorigenesis, which can lead to novel therapeutic applications for HCC patients.
Ethical Approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed Consent
Written informed consent was obtained from all individual participants included in the study.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31471271, 31560311), Guangxi Natural Science Fund for Innovation Research Team (2016GXNSFGA38006) and Guangxi Science and Technology Base and Talent Special Fund (AD17195090).
Disclosure
The authors declare that they have no conflicts of interest (including financial and non-financial interests).
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Fu J, Wang H. Precision diagnosis and treatment of liver cancer in China. Cancer Lett. 2018;412:283–288. doi:10.1016/j.canlet.2017.10.008
3. Han ZG. Functional genomic studies: insights into the pathogenesis of liver cancer. Annu Rev Genomics Hum Genet. 2012;13:171–205. doi:10.1146/annurev-genom-090711-163752
4. Ceol CJ, Houvras Y, Jane-Valbuena J, et al. The histone methyltransferase SETDB1 is recurrently amplified in melanoma and accelerates its onset. Nature. 2011;471(7339):513–517. doi:10.1038/nature09806
5. Kapoor A, Goldberg MS, Cumberland LK, et al. The histone variant macroH2A suppresses melanoma progression through regulation of CDK8. Nature. 2010;468(7327):1105–1109. doi:10.1038/nature09590
6. Zingg D, Debbache J, Schaefer SM, et al. The epigenetic modifier EZH2 controls melanoma growth and metastasis through silencing of distinct tumour suppressors. Nat Commun. 2015;6:6051. doi:10.1038/ncomms7051
7. Segura MF, Fontanals-Cirera B, Gaziel-Sovran A, et al. BRD4 sustains melanoma proliferation and represents a new target for epigenetic therapy. Cancer Res. 2013;73(20):6264–6276. doi:10.1158/0008-5472.CAN-13-0122-T
8. Bonisch C, Hake SB. Histone H2A variants in nucleosomes and chromatin: more or less stable? Nucleic Acids Res. 2012;40(21):10719–10741. doi:10.1093/nar/gks865
9. Dryhurst D, Ishibashi T, Rose KL, et al. Characterization of the histone H2A.Z-1 and H2A.Z-2 isoforms in vertebrates. BMC Biol. 2009;7:86. doi:10.1186/1741-7007-7-86
10. Matsuda R, Hori T, Kitamura H, Takeuchi K, Fukagawa T, Harata M. Identification and characterization of the two isoforms of the vertebrate H2A.Z histone variant. Nucleic Acids Res. 2010;38(13):4263–4273. doi:10.1093/nar/gkq171
11. Dunn CJ, Sarkar P, Bailey ER, et al. Histone hypervariants H2A.Z.1 and H2A.Z.2 play independent and context-specific roles in neuronal activity-induced transcription of Arc/Arg3.1 and other immediate early genes. eNeuro. 2017;4:4. doi:10.1523/ENEURO.0040-17.2017
12. Faast R, Thonglairoam V, Schulz TC, et al. Histone variant H2A.Z is required for early mammalian development. Curr Biol. 2001;11(15):1183–1187. doi:10.1016/S0960-9822(01)00329-3
13. Hua S, Kallen CB, Dhar R, et al. Genomic analysis of estrogen cascade reveals histone variant H2A.Z associated with breast cancer progression. Mol Syst Biol. 2008;4:188. doi:10.1038/msb.2008.25
14. Baptista T, Graca I, Sousa EJ, et al. Regulation of histone H2A.Z expression is mediated by sirtuin 1 in prostate cancer. Oncotarget. 2013;4(10):1673–1685. doi:10.18632/oncotarget.1237
15. Kim K, Punj V, Choi J, et al. Gene dysregulation by histone variant H2A.Z in bladder cancer. Epigenetics Chromatin. 2013;6(1):34. doi:10.1186/1756-8935-6-34
16. Hsu CC, Shi J, Yuan C, et al. Recognition of histone acetylation by the GAS41 YEATS domain promotes H2A.Z deposition in non-small cell lung cancer. Genes Dev. 2018;32(1):58–69. doi:10.1101/gad.303784.117
17. Vardabasso C, Gaspar-Maia A, Hasson D, et al. Histone variant H2A.Z.2 mediates proliferation and drug sensitivity of malignant melanoma. Mol Cell. 2015;59(1):75–88. doi:10.1016/j.molcel.2015.05.009
18. Yang HD, Kim PJ, Eun JW, et al. Oncogenic potential of histone-variant H2A.Z.1 and its regulatory role in cell cycle and epithelial-mesenchymal transition in liver cancer. Oncotarget. 2016;7(10):11412–11423. doi:10.18632/oncotarget.7194
19. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2015;102(43):15545–15550. doi:10.1073/pnas.0506580102
20. Ku M, Jaffe JD, Koche RP, et al. H2A.Z landscapes and dual modifications in pluripotent and multipotent stem cells underlie complex genome regulatory functions. Genome Biol. 2012;13(10):R85. doi:10.1186/gb-2012-13-10-r85
21. Gretzinger TL, Tyagi M, Fontaine CJ, et al. Fetal alcohol spectrum disorder (FASD) affects the hippocampal levels of histone variant H2A.Z-2. Biochem Cell Biol. 2019;97(4):431–436. doi:10.1139/bcb-2018-0240
22. Rivera-Casas C, Gonzalez-Romero R, Vizoso-Vazquez A, et al. Characterization of mussel H2A.Z.2: a new H2A.Z variant preferentially expressed in germinal tissues from Mytilus. Biochem Cell Biol. 2016;94(5):480–490. doi:10.1139/bcb-2016-0056
23. Hiom K. Coping with DNA double strand breaks. DNA Repair (Amst). 2010;9(12):1256–1263. doi:10.1016/j.dnarep.2010.09.018
24. Grabarz A, Barascu A, Guirouilh-Barbat J, Lopez BS. Initiation of DNA double strand break repair: signaling and single-stranded resection dictate the choice between homologous recombination, non-homologous end-joining and alternative end-joining. Am J Cancer Res. 2012;2(3):249–268.
25. Djebali S, Davis CA, Merkel A, et al. Landscape of transcription in human cells. Nature. 2012;489(7414):101–108. doi:10.1038/nature11233
26. Avin BA, Umbricht CB, Zeiger MA. Human telomerase reverse transcriptase regulation by DNA methylation, transcription factor binding and alternative splicing (Review). Int J Oncol. 2016;49(6):2199–2205. doi:10.3892/ijo.2016.3743
27. Xing S, Li Z, Ma W, et al. DIS3L2 promotes progression of Hepatocellular Carcinoma via hnRNP U-mediated alternative splicing. Cancer Res. 2019;79(19):4923–4936. doi:10.1158/0008-5472.CAN-19-0376
28. Gunady MK, Mount SM, Corrada Bravo H. Yanagi: fast and interpretable segment-based alternative splicing and gene expression analysis. BMC Bioinformatics. 2019;20(1):421. doi:10.1186/s12859-019-2947-6
29. Giaimo BD, Ferrante F, Herchenrother A, Hake SB, Borggrefe T. The histone variant H2A.Z in gene regulation. Epigenetics Chromatin. 2019;12(1):37. doi:10.1186/s13072-019-0274-9
30. Eirin-Lopez JM, Gonzalez-Romero R, Dryhurst D, Ishibashi T, Ausio J. The evolutionary differentiation of two histone H2A.Z variants in chordates (H2A.Z-1 and H2A.Z-2) is mediated by a stepwise mutation process that affects three amino acid residues. BMC Evol Biol. 2009;9:31. doi:10.1186/1471-2148-9-31
31. Greenberg RS, Long HK, Swigut T, Wysocka J. Single amino acid change underlies distinct roles of H2A.Z subtypes in human syndrome. Cell. 2019;178(6):1421–1436 e1424. doi:10.1016/j.cell.2019.08.002
32. Chang M, Sun L, Liu X, Sun W, You X. Association of common variants in H2AFZ gene with schizophrenia and cognitive function in patients with schizophrenia. J Hum Genet. 2015;60(10):619–624. doi:10.1038/jhg.2015.89
33. Rangasamy D, Greaves I, Tremethick DJ. RNA interference demonstrates a novel role for H2A.Z in chromosome segregation. Nat Struct Mol Biol. 2004;11(7):650–655. doi:10.1038/nsmb786
34. Chen X, Bhadauria V, Ma B. ChIP-Seq: a powerful tool for studying protein-DNA interactions in plants. Curr Issues Mol Biol. 2018;27:171–180. doi:10.21775/cimb.027.171
35. Lloyd SM, Bao X. Pinpointing the genomic localizations of chromatin-associated proteins: the yesterday, today, and tomorrow of ChIP-seq. Curr Protocols Cell Biol. 2019;84(1):e89. doi:10.1002/cpcb.89
36. Nishibuchi I, Suzuki H, Kinomura A, et al. Reorganization of damaged chromatin by the exchange of histone variant H2A.Z-2. Int J Radiat Oncol Biol Phys. 2014;89(4):736–744. doi:10.1016/j.ijrobp.2014.03.031
37. Nissen KE, Homer CM, Ryan CJ, et al. The histone variant H2A.Z promotes splicing of weak introns. Genes Dev. 2017;31(7):688–701. doi:10.1101/gad.295287.116
38. Jimeno-Gonzalez S, Payan-Bravo L, Munoz-Cabello AM, et al. Defective histone supply causes changes in RNA polymerase II elongation rate and cotranscriptional pre-mRNA splicing. Proc Natl Acad Sci U S A. 2015;112(48):14840–14845. doi:10.1073/pnas.1506760112
39. Soboleva TA, Parker BJ, Nekrasov M, et al. A new link between transcriptional initiation and pre-mRNA splicing: the RNA binding histone variant H2A.B. PLoS Genet. 2017;13(2):e1006633. doi:10.1371/journal.pgen.1006633
40. Neves LT, Douglass S, Spreafico R, Venkataramanan S, Kress TL, Johnson TL. The histone variant H2A.Z promotes efficient cotranscriptional splicing in S. cerevisiae. Genes Dev. 2017;31(7):702–717. doi:10.1101/gad.295188.116
41. Giaimo BD, Ferrante F, Vallejo DM, et al. Histone variant H2A.Z deposition and acetylation directs the canonical Notch signaling response. Nucleic Acids Res. 2018;46(16):8197–8215. doi:10.1093/nar/gky551
42. Hornbeck PV, Zhang B, Murray B, Kornhauser JM, Latham V, Skrzypek E. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 2015;43(Database issue):D512–520. doi:10.1093/nar/gku1267
43. Tsai CH, Chen YJ, Yu CJ, et al. SMYD3-mediated H2A.Z.1 methylation promotes cell cycle and cancer proliferation. Cancer Res. 2016;76(20):6043–6053. doi:10.1158/0008-5472.CAN-16-0500
44. Lumpkin RJ, Gu H, Zhu Y, et al. Site-specific identification and quantitation of endogenous SUMO modifications under native conditions. Nat Commun. 2017;8(1):1171. doi:10.1038/s41467-017-01271-3
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.