Back to Journals » Infection and Drug Resistance » Volume 15
Virulence Genotype and Correlation of Clinical Severeness with Presence of the Type VI Secretion System in Klebsiella pneumoniae Isolates Causing Bloodstream Infections
Authors Zhang Y , Xu Y , Huang Y
Received 20 December 2021
Accepted for publication 26 March 2022
Published 5 April 2022 Volume 2022:15 Pages 1487—1497
DOI https://doi.org/10.2147/IDR.S353858
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Yin Zhang, Yuanhong Xu, Ying Huang
Department of Clinical Laboratory, The First Affiliated Hospital of Anhui Medical University, Hefei, People’s Republic of China
Correspondence: Ying Huang; Yuanhong Xu, Department of Clinical Laboratory, The First Affiliated Hospital of Anhui Medical University, Hefei, People’s Republic of China, Email [email protected]; [email protected]
Background: Klebsiella pneumoniae (K. pneumoniae) causes bloodstream infection (BSI), which is responsible for a high rate of morbidity and mortality among different populations. In mainland China, data on the correlation and features of the type VI secretion system (T6SS) gene cluster in K. pneumoniae is currently scarce. As a result, we conducted a prospective investigation to determine the involvement of the T6SS in K. pneumoniae pathogenicity and antibiotic resistance.
Methods: In this prospective analysis, we enrolled 119 individuals who had been diagnosed with K. pneumoniae bloodstream infection between July 2019 and January 2021 and acquired demographic and clinical data from their medical records. The virulence genes rmpA, rmpA2, aerobactin, iroB, hcp, vgrG, and icmF were tested for K1 and K2, antimicrobial susceptibility. Five T6SS-positive and five T6SS-negative isolates were chosen for the competition, serum resistance, and biofilm formation experiments to further gain insights regarding the microbiological properties of T6SS-positive K. pneumoniae isolates.
Results: Among 119 isolates obtained from patients with BSIs, 20 (16.8%) were T6SS positive K. pneumoniae. T6SS positive strains had four virulence genes and a greater K1 capsular serotypes rate than T6SS negative bacteria. Among hvKP isolates, the T6SS positive rate was substantially greater than the T6SS negative rate (P = 0.001). T6SS-positive K. pneumoniae strains had a lower rate of antimicrobial resistance in comparison to T6SS-negative bacteria. T6SS-positive isolates may be more competitive with Escherichia coli than T6SS-negative isolates. T6SS-positive isolates, on the other hand, did not show stronger biofilm-forming activity or a higher survival rate in the presence of normal human serum in comparison to T6SS-negative isolates.
Conclusion: T6SS-positive K. pneumoniae was common in people who had BSIs. In T6SS‐containing K. pneumoniae, the system may play a major role in bacterial competition.
Keywords: hypervirulent Klebsiella pneumoniae, bloodstream infection, virulence, antibacterial, bacterial competition, type VI secretion system
Introduction
Klebsiella pneumoniae (K. pneumoniae), an opportunistic infecting microbe, frequently causes pneumonia, bacteremia, liver abscess, and urinary tract infection.1,2 Multidrug-resistant strains of K. pneumoniae are common, thus leading to failure of antibiotics therapy against different infections. Although most medicines are effective against nonclassical hypervirulent K. pneumoniae strains, they can induce meddlesome infections in both immunocompromised and healthy patients within the community.3 New K. pneumoniae strains with hypervirulence and multidrug resistance have recently been discovered in China, raising worries about K. pneumoniae infections.4–6
Klebsiella pneumoniae is a common nosocomial pathogen that is causing widespread concern due to multidrug resistance and the recent emergence of hypervirulent strains in clinical settings. Capsule, Pili, lipopolysaccharide (LPS), and iron carriers are now known to be pathogenic factors.7 RmpA is a plasmid-based virulence element in K. pneumoniae that regulates capsular polysaccharide production.8 Enterobactin, yersiniabactin, salmochelin, and aerobactin are the four iron-absorbing molecules (iron carriers) found in K. pneumoniae.9 Aerobactin is thought to be the primary virulence factor of hvKP since it is involved in iron transport, proliferation, and virulence in K. pneumoniae.10 String test in combination with aerobactin and the rmpA index improved the detection rate of hvKP, according to Li et al.11
The bacterial Type VI Secretion System (T6SS) is a membrane-attached contractile phage tail that is physically and mechanistically similar to a membrane-linked intracellular contractile phage tail.12,13 T6SS spike and tube elements, as well as anti-bacterial and anti-eukaryotic effectors, are propelled out of predatory T6SS positive cells and into target cells by a fast conformational shift in the structural framework of a sheath protein complex, according to recent research.14
A functioning T6SS secretes hemolysin-coregulated protein (Hcp) into the extracellular environment.15,16 Owing to their reciprocal needs for secretion, Hcp and another widely reported extracellular protein (or exoprotein), valine-glycine repeat protein G (VgrG), are devoid of a detectable Tat or Sec-dependent signal peptide and are a fundamental element of the secretion apparatus.17 In vitro, Hcp creates a hexamer ring having a 40-pore internal pore and has the ability to be stacked as a tube-like structural form. Surprisingly, the structural similarity and sequence of Hcp and VgrG to T4 bacteriophage spike gp5/gp27 and tail tube gp19 has led to suggesting a Hcp-VgrG phage tail-like structure.18
ATPases or proton motive forces are commonly used in bacterial secretion systems to energize secretion apparatus assembly and/or substrate transfer. A pair of proteins whose ATPase activity or nucleotide-binding sites are already known and could be used as energizers in T6SSs.19 TssH (ClpV), a AAA+-type ATPase, facilitates ATP hydrolysis-mediated remodeling of cogwheel-like TssB/TssC (VipA/VipB) tubules, required for Hcp secretion.12,20 TssM belongs to the intracellular multiplication protein F (IcmF) family and is a T6SS inner membrane protein. TssM’s NTPase activity and role in T6SS remain unknown, apart from the persistence of its Walker A nucleotide-binding motif.21 TssM, belonging to the IcmF family of proteins, has ATPase activity and induces the secretion of type VI, according to one study.18
T6SSs are implicated in anti-virulence or virulence, cytotoxicity to prokaryotic hosts or eukaryotic, and biofilm formation implying that this secretion system has a wide range of functions.22–24 The T6SS gene cluster of Ralstonia solanacearum, for example, plays a key role not only in plant pathogenicity but also in motility and biofilm generation. MFE01tssC, a mutant of Pseudomonas fluorescens MFE01 that lacks one of the primary constituent genes, was not able to form its own biofilm.23 The T6SS has also been linked to biofilm development in Burkholderia cenocepacia.25 This mechanism is also engaged in cell-to-cell communication and signaling. The T6SS is used by Vibrio cholerae to cause variation in the cellular activity of the host that lowers the population of competing bacteria.26,27
The goal of this study was to look at the correlation of the type VI secretion system (T6SS) in K. pneumoniae pathogenicity and antibiotic resistance. T6SS-positive and T6SS-negative K. pneumoniae clinical isolates were examined in terms of antimicrobial resistance and virulence characteristics. In vitro tests, for instance, competition, biofilm formation, and serum resistance assays were used to evaluate T6SS function.
Materials and Methods
Study Designs and Data Collection
The First Affiliated Hospital of Anhui Medical University, a 3000-bed tertiary-level health-care facility in Hefei, China, hosted this single-center prospective cohort study from July 2019 to January 2021. The current investigation was duly approved by the human ethics council of Anhui Medical University’s First Affiliated Hospital. As no personal information was used in the study, patients were not required to provide a declaration of permission for submission.
The BacT/ALERT 3D system was used for culturing K. pneumoniae BSI, which was isolated from one or more sets of aseptically acquired blood culture bottles (Becton-Dickinson, Sparks, MD, USA). Blood was cultured in the microbiology lab employing a BacT/ALERT 3D system (Becton-Dickinson, Sparks, MD, USA). Matrix-associated laser desorption ionization-time of flight mass spectrometry (Vitek MS, Biomerieux, France) was utilized for the recognition of all isolates. Following the 2019 Clinical and Laboratory Standards Institute’s recommendations,28 antibiotic susceptibility testing was done making use of the VITEK 2 (Card number: AST-GN13) system or the methods of Kirby-Bauer Disk Diffusion (Oxoid, UK) (CLSI). Escherichia coli ATCC25922 and K. pneumoniae ATCC700603 were employed as quality control isolates. ESBL was confirmed by agar dilution test using ceftazidime and cefotaxime combined with clavulanate.29 Carbapenem (ertapenem, meropenem and/or imipenem) resistance was confirmed by disk diffusion method.30
Clinical Data and Definitions
Medical data were used to acquire clinical information on 119 K. pneumoniae-infected patients. Immunosuppression was referred to as primary immunodeficiency disorder and/or regular use of high-dose steroid therapy (prednisolone 10 mg/daily or comparable dose) for more than half a month, and/or immunosuppressive chemotherapy within the previous two months. Neutropenia was classified as a neutrophil count of fewer than 500 per liter. When symptoms and positive blood culture were gathered more than 48 hours following hospital admission, it was considered a nosocomial infection. MDR (multi-drug resistance) is referred to as acquired non-susceptibility of at least one antimicrobial entity from three or more groups. The length of duration between the automated system’s alert signal and the start of blood incubation and was assessed as time to positive (TTP).31 In case of the presence of more than one positive sample, we used the shortest TTP. Appropriate antibiotic therapy meant receiving at least one intravenous active antimicrobial treatment based on susceptibility results within 24 hours of collecting blood samples and before susceptibility findings were available; in any other circumstance, unsuitable antimicrobial therapy was defined.32
Detection of Virulence Genes, Capsular Serotypes, and T6SS Genes
A commercial DNA extraction kit was used to extract DNA from K. pneumoniae isolates (Sangon Biotech, Shanghai, China). From an overnight culture, 1mL of bacterial suspension matched comparable to 0.5 McFarland was made and centrifuged. DNA was extracted from the pellet according to the kit’s instructions and then resolved in 100µL TE buffer.
PCR-based detection of biomarker genes employed 12.5 μL of 2× Spark Taq PCR Master Mix (Solely Bio, Shandong, China), 1 μL of forward primer, 1 μL of reverse primer (10 pmol/μL primer stock), 1 μL of genomic DNA, and 9.5 μL of water. PCR was performed using a Biometra thermocycler with the cycling conditions mentioned as follows: First step: 94.0°C for 3 min; Second step: 94.0°C for 30s; Third step: primer-specific annealing temperature for 30s; Forth step: 72°C for 1 min; Fifth step: repeating steps 2–4 for 35 cycles; Sixth step: 72.0°C for 10 min; Seventh step: holding at 4°C. 1% agarose gel was used to resolve the amplified products. Table S1 lists all the primers employed.
Biofilm Assay
Biofilm formation tests were carried out as described earlier, with a number of tweaks.33 The OD600 was used to standardize overnight cultures. The suspension was inoculated into the wells of a polystyrene 96-well flat-bottom plate using a 200-μL aliquot and left to incubate for 18 hours at 37°C. The medium was then taken from the plates, and each well was thoroughly washed three times with water. After 30 minutes of air-drying, crystal violet solution (1%, 150 μL) was applied to individual wells for 15 minutes. The colorant was discarded after staining the adherent cells, and 200 μL of distilled water was used to rinse the wells three times in order to eliminate any surplus stain. For 0.5 hours, the plates were air-dried. In 200 μL of 100% ethanol, the dye absorbed by the adhering cells was solubilized. As a negative control, sterile LB was employed. The stained adherent bacteria and control wells had their OD600 values tested. Three separate studies, each with three replicate wells, were carried out. Biofilm production positive was defined as a strain with an absorbance value greater than 2 times that of the blank absorbance.
Serum Resistance Assay
The serum resistance analyses were carried out as elaborated earlier, with minimal modifications.34 Dilution 1:100 of overnight bacterial cultures was carried out into fresh LB media (10 mL) and the diluted specimens were allowed to incubate until an OD600 of 0.5 was achieved. The culture was then rinsed in phosphate-buffered saline (PBS) for 1 mL followed by resuspension in 1 mL of PBS. Following that, 100 μL of the bacterial suspension were combined with 300 μL of normal human serum (NHS). The suspensions of serum-bacteria were mixed and then set to incubate at 37°C for 3 hours. A 100-µL aliquot of each culture was obtained prior to and following the 3-h period of incubation, diluted serially, and plated for calculating the serum bactericidal effect. The serum bactericidal effect was calculated using the ratio of serum-bacteria suspension CFUs to bacterial suspension CFUs without NHS. All studies were carried out in triplicate, and the findings are expressed as percent survival.
In vitro Competition Assay
Strains of the attacker (K. pneumoniae) and prey (E. coli MG1655) were cultured in LB overnight at 37°C. Each culture was then diluted 1:100 in 10 mL of LB broth after being standardized to an OD600 of 0.5. The suspensions of bacteria were mixed in a 2:1 attacker: prey ratio and cultured for 20 hours at 180 rpm and 37°C. The determination of the cell count for each strain was carried out via pouring their serial 10-fold dilutions onto LB agar plates in the presence or absence of ampicillin (50 mg/L).25,35 An in vitro competitive index was calculated to measure the relative fitness of K. pneumoniae isolates against E. coli strain (CI). The CI was calculated as the ratio of ampicillin-resistant K. pneumoniae to ampicillin-susceptible CFUs E. coli. A total of ten competition experiments were carried out.
Statistical Analysis
In the case of continuous variables, medians and interquartile ranges (IQR) were used, whereas, for categorical variables, relative and absolute frequencies were employed. The Student’s t-test or the Mann–Whitney U-test, as well as the 2 test or Fisher’s exact test, were used for the comparison of categorical and continuous parameters. All statistical tests were two-tailed, and statistical significance was referred to as a P-value equivalent to 0.05. The SPSS statistical package v.24.0 (SPSS Inc., Chicago, IL, USA) was employed to conduct the analyses, and Prism software v.9.0 (GraphPad Inc., La Jolla, CA, USA) was used to create the visualizations.
Results
Study Population and Patient Characteristics
In the duration of the study, one hundred and thirty inpatients with a positive K. pneumoniae blood culture were included. Eleven of them were disqualified (five cases were newborns, four cases were infected with other bacteria, two cases had incomplete information). As a result, 119 instances were finally examined in this work.
The average age of these individuals was 51.6±16.9 years, with men accounting for 69.7% (83/119) of the total. The average duration of the stay in the hospital was 37.9±45.2 days. Hypoalbuminemia (82.3%, 98/119) was the most frequent underlying condition, followed by immunosuppression (43.7%, 52/119). Diabetes (32/119, 26.9%) was the most common underlying condition, followed by hypertension and end-stage renal disease (22/119, 18.4%). Forty-seven patients (or 39.4%) were admitted to the ICU. Forty patients (33.6%) were community-acquired infection. The Charlson Comorbidity Index has a median of 1.8 (0–6.00). Eighty-three patients (69.7%) received suitable empirical antibiotic treatment. MDR microorganisms were found in seventy-eight (65.5%) of the patients (78/119). The mortality rate at the hospital was 36.1% (43/119). Table 1 provides more information on clinical features.
 |
Table 1 Baseline Characteristics of Patients with Klebsiella pneumoniae Bloodstream Infection |
The features of the positive and negative T6SS groups are depicted in Table 1. Patients in the T6SS negative group had significantly greater 30-day mortality (37.4% vs 10%, P = 0.01), MDR (71.7% vs 35.0%, P = 0.002), and immunosuppressive patients (48.5% vs 20.0%, P = 0.01). T6SS positive patients had higher rates of hypertension than T6SS negative patients (40.0% vs 14.1%, P = 0.007). There were no considerable differences in the underlying diseases, community infectious infection, antibiotic administration, or length of hospitalized stay.
Microbiological Characteristics and Antimicrobial Susceptibility
Apart from natural resistance to ampicillin, K. pneumoniae had the highest rate of resistance towards piperacillin, whereas tigecycline resistance was as low as 0.8%. T6SS-positive K. pneumoniae strains also had lower rates of antimicrobial resistance than T6SS-negative bacteria. With the exception of minocycline, tigecycline, and ceftazidime avibactam, practically all antibiotic drugs had significantly greater resistance rates for T6SS negatives than for T6SS positives. Twenty-four (20.2%) of the 119 K. pneumoniae clinical isolates were found to be ESBL-producing. Seventy-eight strains (65.5%) were found to be multidrug-resistant bacteria (MDR), which were shown to be more prevalent in the T6SS-negative group (71.7% vs 35.0%, P = 0.002). Carbapenem-resistant K. pneumoniae (CRKP) was also found in lower numbers in the T6SS-positive strain (20.0% vs 45.4%, P = 0.03). The most prevalent carbapenemase genes in K. pneumoniae isolates was KPC (n = 43; 87.8%), followed by NDM (n = 3; 6.1%), IMP (n = 1; 2.0%) and VIM (n = 1; 2.0%). Besides, there was no difference in metallo-beta-lactamases harbouring distribution between the two groups (P = 0.594). Table 2 has more information on these findings.
 |
Table 2 Antimicrobial Resistance of T6SS‐positive and T6SS‐negative K. pneumoniae Bloodstream Isolates |
Table 3 shows how T6SS positive and T6SS negative strains differ in terms of capsular serotypes and virulence characteristics. K1 and K2 capsular serotypes accounted for 10.9% (13/119) and 7.5% (9/119) of all K. pneumoniae, respectively. T6SS-positive strains displayed considerably greater prevalences of the K1 serotype (25.0% vs 8.1%, P = 0.027), aerobactin (55.0% vs 17.2%, P0.001), and iroB (45.0% vs 18.2%, P = 0.009) than T6SS-positive strains. T6SS-positive strains had a considerably greater incidence of hvkp than T6SS-negative strains (50.0% vs 16.2%, P = 0.001).
 |
Table 3 Capsular Serotypes and Virulence Factors Between T6SS‐positive and T6SS‐negative K. pneumoniae Bloodstream Isolates |
Crude Impact of TTSS Genotype in Mortality
Table 1 shows that the overall 30-day mortality rate was 32.8% (39 patients), with 21.8% (26 patients) dying within the first seven days of being admitted to the hospital. There were no significant variations in early (7-day) mortality according to the T6SS genotype. Late mortality (30-day) among patients infected with T6SS-positive isolates, on the other hand, was 10% compared to 37.4% among patients infected with T6SS-negative strains (P = 0.01).
Biofilm Formation, Competitive Index, and Serum Resistance
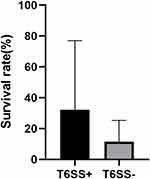
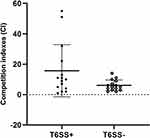
T6SS-positive and T6SS-negative strains had their relative biofilm-forming activity calculated, and individual isolates were investigated in triplicate. T6SS-positive K. pneumoniae isolates’ mean OD600 value was not substantially different from T6SS-negative isolates (mean ± standard deviation [SD], 0.059 ± 0.005 vs 0.08 ± 0.04; P = 0.303) (Figure 1). After 3 h of incubation in serum, T6SS-positive isolates revealed no significant changes in survival rate compared to T6SS-negative isolates (40.95% vs 21.73%; P = 0.35) (Figure 2). In vitro competition experiments were also carried out to calculate the CI to see if T6SS-positive isolates could outcompete other bacteria. This study calculates the CI of 5 T6SS positive and 5 T6SS negative K. pneumoniae, as shown in Figure 3. The average CI of T6SS positive K. pneumoniae was 15.67 and that of T6SS negative K. pneumoniae was 6.06. The competitiveness of T6SS positive strains was higher than that of T6SS negative strains (t = 2.117, P < 0.05). The result has shown that T6SS-positive isolates were more competitive during joint incubation than T6SS-negative isolates (Figure 3).
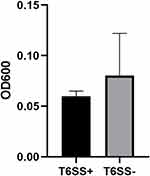 |
Figure 1 Mean ± standard deviation optical density at 600 nm of T6SS-positive and T6SS-negative Klebsiella pneumoniae isolates stained with crystal violet in the biofilm formation assay. |
Discussion
In this investigation, strains positive for rmpA and aerobactin were classified as hvKP, and the results revealed that 26 (21.8%) of the strains were hvKP. T6SS-positive strains were defined as those that were positive for icmF, vgrG, and hcp in a prior investigation. Based on these criteria, our research found that T6SS genes were present in 16.8% of K. pneumoniae bloodstream isolates. However, only patients with bloodstream infections were included in this investigation, not individuals with chronic infections who might have been colonized by mature K. pneumoniae colonies. T6SS positive strains had a greater detection rate of K1 capsular serotypes and four virulence genes than T6SS negative bacteria, according to the findings. The incidence of hvkp was substantially greater in T6SS-positive strains in comparison to the T6SS-negative strains. This research backed up the pathogenicity of the T6SS-positive K. pneumoniae strains.
T6SS-positive K. pneumoniae strains had lower antimicrobial resistance rates than T6SS-negative bacteria in this investigation. With the exception of minocycline, tigecycline, and ceftazidime avibactam, practically all antibiotic drugs had significantly greater resistance rates for T6SS negatives than for T6SS positives, which may be related to the distribution of metallo-beta-lactamases.36 The percentage of multidrug-resistant, carbapenem-resistant, and ESBL-producing K. pneumoniae, on the other hand, was extremely high. Multidrug resistance levels linked with this species have skyrocketed, posing a global threat, in particular for carbapenemase-producing K. pneumoniae.37–39 The drug-resistance pattern of T6SS in K. pneumoniae, on the other hand, is little understood. A study found that lactam antibiotics, at a sub-inhibitory concentration, increased T6SS-dependent killing by stimulating the production and secretion of the CRKP HS11286 T6SS.40 As a result of the antibiotic stress, CRKP HS1186 outperforms the T6SS/multidrug-resistant strain in terms of growth superiority.
T6SS-positive isolates had stronger biofilm-forming activity and survived longer when present in normal human serum than T6SS-negative isolates, according to previous research. However, the findings of our study on K. pneumoniae isolates contradict this concept. The differences between K. pneumoniae and other bacteria could be due to species-specific characteristics. It is unclear whether T6SS-positive isolates’ biofilm-forming ability and serum resistance are related to the T6SS’s own function. A number of investigations have shown that the T6SS is involved in biofilm production in a variety of bacteria; however, certain bacteria do not need the T6SS to create biofilms. Type 3 pili and capsular polysaccharides (CPs) are the most critical surface features of K. pneumoniae that take part in the production process.41,42 Pili mediate long-term adherence, whereas CPs influence intercellular communication and biofilm structure in the end. The function of biofilm can also be affected by mutations in certain K. pneumoniae genes.43
In this investigation, we discovered that T6SS-positive isolates outperformed T6SS-negative isolates in terms of outcompeting E. coli. In a vast majority of T6SS-carrying bacteria, the significance of the T6SS in intraspecies and interspecies bacterial competition has been recognized as a key characteristic of the system. The T6SS has been shown to direct effector proteins to other bacteria of the likes of P. aeruginosa, V. cholerae, and S. marcescens, allowing the organism to get involved in competition more successfully against other bacteria in its growing environment.22,25,44,45 Nevertheless, the antibacterial properties of the T6SS in K. pneumoniae have yet to be discovered. The T6SS renders a fitness advantage for T6SS-positive clinical isolates of K. pneumoniae, according to our findings.
Conclusion
T6SS-positive K. pneumoniaeThe prevalence of T6SS-positive K. pneumoniae was 16.8% in people who had BSIs. T6SS-positive K. pneumoniae strains had a lower rate of antimicrobial resistance in comparison to T6SS-negative strains. T6SS-positive isolates may be more competitive with Escherichia coli than T6SS-negative isolates. T6SS-positive isolates, on the other hand, did not show stronger biofilm-forming activity or a higher survival rate in the presence of normal human serum in comparison to T6SS-negative isolates.
Abbreviations
T6SS, type VI secretion system; BSI, bloodstream infection; hvKp, hypervirulent Klebsiella pneumoniae; Hcp, hemolysin-coregulated protein; VgrG, valine-glycine repeat protein G; IcmF, iintracellular multiplication protein F; TssM, T6SS inner membrane protein; MDR, multi-drug resistance; TTP, time to positive; PBS, phosphate-buffered saline; NHS, normal human serum; CFUs, colony forming units; IQR, interquartile ranges; CCI, Charlson Comorbidity Index; CRKP, carbapenem-resistant Klebsiella pneumoniae; LPS, lipopolysaccharide.
Data Sharing Statement
All data generated or analyzed during this study are included in this manuscript.
Ethics Approval and Consent to Participate
This study conformed to the Declaration of Helsinki and was approved by the Medical Ethics Committee of the First Affiliated Hospital of Anhui Medical University. In this study, informed consent was waived by using anonymous clinical data.
Acknowledgments
We thank the First Affiliated Hospital of Anhui Medical University for their cooperation.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
The work was supported by National Natural Science Foundation of China (No.:81171606).
Disclosure
The authors declare that they have no conflicts of interest in this work.
References
1. Zhou C, Wu Q, He L, et al. Clinical and molecular characteristics of carbapenem-resistant hypervirulent Klebsiella pneumoniae isolates in a tertiary hospital in Shanghai, China. Infect Drug Resist. 2021;14:2697–2706. doi:10.2147/IDR.S321704
2. Ejaz H, Wang N, Wilksch JJ, et al. Phylogenetic analysis of Klebsiella pneumoniae from Hospitalized Children, Pakistan. Emerg Infect Dis. 2017;23(11):1872–1875. doi:10.3201/eid2311.170833
3. Heinz E, Ejaz H, Bartholdson Scott J, et al. Resistance mechanisms and population structure of highly drug resistant Klebsiella in Pakistan during the introduction of the carbapenemase NDM-1. Sci Rep. 2019;9(1):2392. doi:10.1038/s41598-019-38943-7
4. Russo TA, Marr CM. Hypervirulent Klebsiella pneumoniae. Clin Microbiol Rev. 2019;32(3):e00001–19. doi:10.1128/CMR.00001-19
5. Wang X, Xie Y, Li G, et al. Whole-genome-sequencing characterization of bloodstream infection-causing hypervirulent Klebsiella pneumoniae of capsular serotype K2 and ST374. Virulence. 2018;9(1):510–521. doi:10.1080/21505594.2017.1421894
6. Liu C, Guo J. Hypervirulent Klebsiella pneumoniae (hypermucoviscous and aerobactin positive) infection over 6 years in the elderly in China: antimicrobial resistance patterns, molecular epidemiology and risk factor. Ann Clin Microbiol Antimicrob. 2019;18(1):4. doi:10.1186/s12941-018-0302-9
7. Bei L, Zhao Y, Liu C, Chen Z, Zhou D. Molecular pathogenesis of Klebsiella pneumoniae. Future Microbiol. 2014;9(9):1071–1081. doi:10.2217/fmb.14.48
8. Hsu CR, Lin TL, Chen YC, Chou HC, Wang JT. The role of Klebsiella pneumoniae rmpA in capsular polysaccharide synthesis and virulence revisited. Microbiology. 2011;157(Pt 12):3446–3457. doi:10.1099/mic.0.050336-0
9. Russo TA, Shon AS, Beanan JM, et al. Hypervirulent K. pneumoniae secretes more and more active iron-acquisition molecules than ‘classical’ K. pneumoniae thereby enhancing its virulence. PLoS One. 2011;6(10):e26734. doi:10.1371/journal.pone.0026734
10. Russo TA, Olson R, Fang CT, et al. Identification of biomarkers for differentiation of hypervirulent Klebsiella pneumoniae from classical K. pneumoniae. J Clin Microbiol. 2018;56(9):e00776–18. doi:10.1128/JCM.00776-18
11. Li J, Ren J, Wang W, et al. Risk factors and clinical outcomes of hypervirulent Klebsiella pneumoniae induced bloodstream infections. Eur J Clin Microbiol Infect Dis. 2018;37(4):679–689. doi:10.1007/s10096-017-3160-z
12. Basler M, Pilhofer M, Henderson GP, Jensen GJ, Mekalanos JJ. Type VI secretion requires a dynamic contractile phage tail-like structure. Nature. 2012;483(7388):182–186. doi:10.1038/nature10846
13. Ho BT, Dong TG, Mekalanos JJ. A view to a kill: the bacterial type VI secretion system. Cell Host Microbe. 2014;15(1):9–21. doi:10.1016/j.chom.2013.11.008
14. Kudryashev M, Wang RY, Brackmann M, et al. Structure of the type VI secretion system contractile sheath. Cell. 2015;160(5):952–962. doi:10.1016/j.cell.2015.01.037
15. Silverman JM, Agnello DM, Zheng H, et al. Haemolysin coregulated protein is an exported receptor and chaperone of type VI secretion substrates. Mol Cell. 2013;51(5):584–593. doi:10.1016/j.molcel.2013.07.025
16. Pukatzki S, Ma AT, Sturtevant D, et al. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc Natl Acad Sci USA. 2006;103(5):1528–1533. doi:10.1073/pnas.0510322103
17. Monjarás Feria J, Valvano MA. An overview of anti-eukaryotic T6SS effectors. Front Cell Infect Microbiol. 2020;10:584751. doi:10.3389/fcimb.2020.584751
18. Ma LS, Narberhaus F, Lai EM. IcmF family protein TssM exhibits ATPase activity and energizes type VI secretion. J Biol Chem. 2012;287(19):15610–15621. doi:10.1074/jbc.M111.301630
19. Bönemann G, Pietrosiuk A, Diemand A, Zentgraf H, Mogk A. Remodelling of VipA/VipB tubules by ClpV-mediated threading is crucial for type VI protein secretion. EMBO J. 2009;28(4):315–325. doi:10.1038/emboj.2008.269
20. Pietrosiuk A, Lenherr ED, Falk S, et al. Molecular basis for the unique role of the AAA+ chaperone ClpV in type VI protein secretion. J Biol Chem. 2011;286(34):30010–30021. doi:10.1074/jbc.M111.253377
21. Ma LS, Lin JS, Lai EM. An Icm F family protein, Imp LM, is an integral inner membrane protein interacting with Imp KL, and its walker a motif is required for type VI secretion system-mediated Hcp secretion in Agrobacterium tumefaciens. J Bacteriol. 2009;191(13):4316–4329. doi:10.1128/JB.00029-09
22. Murdoch SL, Trunk K, English G, Fritsch MJ, Pourkarimi E, Coulthurst SJ. The opportunistic pathogen Serratia marcescens utilizes type VI secretion to target bacterial competitors. J Bacteriol. 2011;193(21):6057–6069. doi:10.1128/JB.05671-11
23. Gallique M, Decoin V, Barbey C, et al. Contribution of the Pseudomonas fluorescens MFE01 type VI secretion system to biofilm formation. PLoS One. 2017;12(1):e0170770. doi:10.1371/journal.pone.0170770
24. Decoin V, Gallique M, Barbey C, et al. A Pseudomonas fluorescens type 6 secretion system is related to mucoidy, motility and bacterial competition. BMC Microbiol. 2015;15:72. doi:10.1186/s12866-015-0405-9
25. Spiewak HL, Shastri S, Zhang L, et al. Burkholderia cenocepacia utilizes a type VI secretion system for bacterial competition. Microbiologyopen. 2019;8(7):e774. doi:10.1002/mbo3.774
26. Joshi A, Kostiuk B, Rogers A, Teschler J, Pukatzki S, Yildiz FH. Rules of engagement: the type VI secretion system in Vibrio cholerae. Trends Microbiol. 2017;25(4):267–279. doi:10.1016/j.tim.2016.12.003
27. Manera K, Caro F, Li H, et al. Sensing of intracellular Hcp levels controls T6SS expression in Vibrio cholerae. Proc Natl Acad Sci USA. 2021;118(25):e2104813118. doi:10.1073/pnas.2104813118
28. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing. CLSI Supplement M100.
29. Ejaz H, Younas S, Qamar MU, et al. Molecular epidemiology of extensively drug-resistant mcr encoded colistin-resistant bacterial strains co-expressing multifarious β-lactamases. Antibiotics. 2021;10(4):467. doi:10.3390/antibiotics10040467
30. Zhang X, Li F, Cui S, et al. Prevalence and distribution characteristics of blaKPC-2 and blaNDM-1 genes in Klebsiella pneumoniae. Infect Drug Resist. 2020;13:2901–2910. doi:10.2147/IDR.S253631
31. Ning Y, Hu R, Yao G, Bo S. Time to positivity of blood culture and its prognostic value in bloodstream infection. Eur J Clin Microbiol Infect Dis. 2016;35(4):619–624. doi:10.1007/s10096-016-2580-5
32. Cunha BA. Sepsis and septic shock: selection of empiric antimicrobial therapy. Crit Care Clin. 2008;24(2):313–34, ix. doi:10.1016/j.ccc.2007.12.015
33. Merritt JH, Kadouri DE, O’Toole GA. Growing and analyzing static biofilms. Curr Protoc Microbiol. 2005;2(1):1B.
34. Shin J, Ko KS. Effect of plasmids harbouring blaCTX-M on the virulence and fitness of Escherichia coli ST131 isolates. Int J Antimicrob Agents. 2015;46(2):214–218. doi:10.1016/j.ijantimicag.2015.04.012
35. Weber BS, Miyata ST, Iwashkiw JA, et al. Genomic and functional analysis of the type VI secretion system in Acinetobacter. PLoS One. 2013;8(1):e55142. doi:10.1371/journal.pone.0055142
36. Shirley M. Ceftazidime-avibactam: a review in the treatment of serious gram-negative bacterial infections. Drugs. 2018;78(6):675–692. doi:10.1007/s40265-018-0902-x
37. Imtiaz W, Syed Z, Rafaque Z, Andrews SC, Dasti JI. Analysis of antibiotic resistance and virulence traits (genetic and phenotypic) in Klebsiella pneumoniae clinical isolates from Pakistan: identification of significant levels of carbapenem and colistin resistance. Infect Drug Resist. 2021;14:227–236. doi:10.2147/IDR.S293290
38. Ding Y, Wang H, Pu S, Huang S, Niu S. Resistance trends of Klebsiella pneumoniae causing urinary tract infections in Chongqing, 2011–2019. Infect Drug Resist. 2021;14:475–481. doi:10.2147/IDR.S295870
39. Qamar MU, Ejaz H, Walsh TR, et al. Clonal relatedness and plasmid profiling of extensively drug-resistant New Delhi metallo-β-lactamase-producing Klebsiella pneumoniae clinical isolates. Future Microbiol. 2021;16:229–239. doi:10.2217/fmb-2020-0315
40. Liu L, Ye M, Li X, et al. Identification and characterization of an antibacterial type VI secretion system in the Carbapenem-resistant strain Klebsiella pneumoniae HS11286. Front Cell Infect Microbiol. 2017;7:442. doi:10.3389/fcimb.2017.00442
41. De oliveira Júnior NG, Franco OL. Promising strategies for future treatment of Klebsiella pneumoniae biofilms. Future Microbiol. 2020;15:63–79. doi:10.2217/fmb-2019-0180
42. Clegg S, Murphy CN, Mulvey MA, Stapleton AE, Klumpp DJ. Epidemiology and virulence of Klebsiella pneumoniae. Microbiol Spectr. 2016;4(1). doi:10.1128/microbiolspec.UTI-0005-2012
43. Mostafavi M, Wang L, Xie L, et al. Interplay of Klebsiella pneumoniae fabZ and lpxC mutations leads to LpxC inhibitor-dependent growth resulting from loss of membrane homeostasis. mSphere. 2018;3(5):e00508–18. doi:10.1128/mSphere.00508-18
44. Hood RD, Singh P, Hsu F, et al. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe. 2010;7(1):25–37. doi:10.1016/j.chom.2009.12.007
45. MacIntyre DL, Miyata ST, Kitaoka M, Pukatzki S. The Vibrio cholerae type VI secretion system displays antimicrobial properties. Proc Natl Acad Sci USA. 2010;107(45):19520–19524. doi:10.1073/pnas.1012931107
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.