Back to Journals » Infection and Drug Resistance » Volume 12
Virulence Factors Of Carbapenem-Resistant Pseudomonas aeruginosa In Hospital-Acquired Infections In Mansoura, Egypt
Authors El-Mahdy R , El-Kannishy G
Received 8 July 2019
Accepted for publication 24 October 2019
Published 7 November 2019 Volume 2019:12 Pages 3455—3461
DOI https://doi.org/10.2147/IDR.S222329
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Joachim Wink
Rasha El-Mahdy,1 Ghada El-Kannishy2
1Department of Medical Microbiology And Immunology, Faculty of Medicine, Mansoura University, Mansoura, Egypt; 2Department of Internal Medicine, Faculty of Medicine, Mansoura University, Mansoura, Egypt
Correspondence: Rasha El-Mahdy
Faculty of Medicine, Mansoura University, Mansoura 35516, Egypt
Tel +20 5010 0532 9819
Email [email protected]
Purpose: The problem of carbapenem-resistant Pseudomonas aeruginosa in health-care settings is growing worse. This study was conducted to investigate the rate of carbapenemase genes, antibiotic resistance, and virulence factors in carbapenem-resistant P. aeruginosa associated with hospital-acquired infections.
Patients and methods: Isolates of P. aeruginosa were collected from patients with hospital-acquired infections at Mansoura University Hospital in Mansoura. Carbapenem susceptibility was done by broth dilution. The presence of carbapenemase genes and quorum-sensing genes was assessed by PCR. Production of protease, pyocyanin, twitching motility, hemolytic activity and biofilm formation was evaluated.
Results: Out of 80 P. aeruginosa isolates, 34 (42.5%) were resistant to carbapenem. Among carbapenem-resistant P. aeruginosa isolates, 21 (61.8%) were carbapenemase producers. The most prevalent gene detected was blaVIM. The frequency of protease, pyocyanin, twitching motility, hemolytic activity and biofilm formation was 76.2%, 58.8%, 83.8%, 93.8% and 77.5%, respectively. Biofilm formation was significantly associated with carbapenem-resistant P. aeruginosa. On the other hand, pyocyanin production was significantly lower in carbapenem-resistant isolates. No correlation existed between carbapenem resistance and any other studied virulence factors or quorum-sensing genes.
Conclusion: Association of carbapenem-resistant P. aeruginosa with other antibiotic resistance or the presence of virulence factors in hospital-acquired infection may represent a warning that enhances the need for a stringent surveillance program.
Keywords: Pseudomonas aeruginosa, carbapenem, virulence factor, resistance
Introduction
Hospital-acquired multidrug-resistant (MDR) Pseudomonas aeruginosa infections are increasing worldwide and have become a global issue.1 Though carbapenems represent the most effective antibiotics in the treatment of MDR P. aeruginosa infections, carbapenem resistance has been increasingly described all over the world.2 Various mechanisms are involved in carbapenem resistance such as carbapenemase production, intrinsic RND efflux pump systems and lack of outer membrane porin (OprD).3 Carbapenemase genes represent a serious issue, as the resistance can be transmitted horizontally to other species.5 Metallo-β-lactamase (MBL) represents the principal carbapenemases formed by P. aeruginosa.4
Pathogenesis of P. aeruginosa is multifactorial, and many virulence factors are produced that include secreted factors such as alkaline protease, elastase, exotoxin A, pyoverdine, pyocyanin, rhamnolipid structural component lipopolysaccharide, pili, flagella and biofilm formation.6 Production of various virulence factors was regulated by two quorum-sensing (QS) systems las and rhl.7
The association between virulence and resistance in P. aeruginosa is still poorly understood.8 The aim of this study was to determine the occurrence of antibiotic resistance, carbapenemase genes and virulence factors in carbapenem-resistant P. aeruginosa associated with hospital-acquired infections at Mansoura University Hospitals.
Materials And Methods
Study Setting
The study was conducted in Mansoura University Hospital, Mansoura, Egypt. This study was approved by the Mansoura University’s Faculty of Medicine Institutional Review Board—Code number: R.17.10.09.
Isolates
A total nonduplicate 80 isolates of P. aeruginosa isolated from clinical samples from September 2018 to April 2019, recovered from patients with hospital-acquired infections at Mansoura University Hospital.
Isolates of P. aeruginosa were identified as non-lactose fermenting colonies on MacConkey’s medium, Gram’s stain, positive oxidase reaction, citrate utilization test, triple sugar iron, growth at 42°C and growth on cetrimide agar.9 Identification of isolates was confirmed by API 20E (BioMérieux, Marcy l’Étoile, France).
Antimicrobial Susceptibility Testing
The following antibiotics were tested: polymyxin B (300 units), colistin (10 μg), ceftazidime (30 μg), cefepime (30 μg), piperacillin (100μg), piperacillin/tazobactam (100/10 μg), aztreonam (30 μg), tobramycin (10 μg), gentamicin (10 μg), amikacin (30 μg), levofloxacin (10 μg), ciprofloxacin (5 μg), (Oxoid, UK) by disk diffusion method according to the Clinical and Laboratory Standards Institute protocol.10
Minimum inhibitory concentrations (MICs) of imipenem and meropenem (manufacturers: GlaxoSmithKlein, AstraZeneca Pharma, Cairo, Egypt) were tested by broth microdilution method according to Clinical Laboratory Standards Institute guidelines. P. aeruginosa isolates defined as carbapenem resistant when imipenem or/and meropenem MIC ≥8mg/L determined.11
Resistance phenotypes were defined as MDR P. aeruginosa for isolate that is non-susceptible to at least one agent in ≥3 antimicrobial categories; XDR isolate is non-susceptible to at least one agent in all but ≤2categories; pandrug resistant means non-susceptible to all antimicrobial categories.12
Phenotypic Detection Of Carbapenemases
Carbapenem-resistant isolates were screened for carbapenemase production by modified Hodge test (MHT)13 and a combined disk diffusion method.14
Carbapenemases And Quorum-Sensing Genes
PCR for the following carbapenemase genes blaIMP; blaVIM; blaNDM; blaKPC; blaOXA-48 and quorum-sensing genes lasR and rhlR was done as previously described.15,16 Genes used in this study are listed in Table 1.
 |
Table 1 Specific Primers Used In This Study |
Hemolytic Activity
Five microliters of each strain were streaked on agar base supplemented with 5% sheep erythrocytes and then incubated overnight at 22°C and 37°C. Plates were examined for the presence of β-hemolysis around the colonies.17
Twitching Motility
Each strain was stabbed with a sterile toothpick to the bottom 1% Luria–Bertani agar plate and then incubated overnight at 37°C. Twitching motility was evaluated by the presence of a hazy zone surrounding the point of inoculation at the agar–plate interface. Another way to visualize motility was by removal of the agar and addition of crystal violet for 5 mins and then rinsing with tap water.18
Pyocyanin Assay
Culture grew in Pseudomonas broth (20 g peptone, 1.4 g MgCl2 and 10 g K2SO4 per liter of distilled water). Pyocyanin was extracted as previously described.19 Concentrations were determined by multiplying the optical density at 520 nm (OD520) by 17.072. Assays were performed three times. P. aeruginosa PAO1 was used as a positive control strain for pyocyanin assay.
Protease Assay
Strains of P. aeruginosa were streaked on skim milk agar plates (10% w/v skimmed milk) and then incubated at 37°C for 24 hrs. The presence of a clear zone surrounding the colonies indicates a positive test.17
Biofilm
Biofilm formation was evaluated by the semiquantitative determination. Briefly,
125 µL of diluted overnight growth of isolates was inoculated in sterile flat-bottomed 96-well microtiter plates and incubated at 37°C for 24 hrs. Each well was washed three times with 300 µL distilled water, dried in an inverted position at room temperature. 125 µL of 0.1% crystal violet was added for 10–15 mins then rinsed three times with distilled water. 2 mL of 95% ethanol was added. Uninoculated medium was used as control. Spectrophotometer was used to measure the absorbance at 600 nm.20
Statistical Analysis
Data were statistically analyzed using the Statistical Package for Social Sciences (SPSS) version 16 (SPSS Inc, Chicago, IL, USA). Qualitative data are described as numbers and percentages. The Chi-square test or Fisher’s exact test was used for comparison between groups, as appropriate. Results with p<0.05 were considered significant.
Results
Clinical Isolates
In our study, 80 isolates of P. aeruginosa were isolated from different clinical samples, including wound (n = 33), burn (n = 30), respiratory tract (n = 6), urine (n = 6) and blood (n = 5).
Susceptibility Pattern
All isolates were sensitive to colistin, polymyxin B. The susceptibility rate reached up to 52.5%, 45%, 42.5% and 37.5% toward piperacillin/tazobactam, levofloxacin, ciprofloxacin and aztreonam, respectively. Ceftazidime was the least effective antibiotic, with the susceptibility rate being 15%. Fifty-eight (72.5%) isolates were MDR or extensive drug resistance (XDR).
Of the 80 isolates, 34 (42.5%) were carbapenem resistant. Modified Hodge test (MHT) and combined disc diffusion test were positive in 22 (64.7%) and 18 (52.9%), respectively. The sensitivity of MHT and combined disc diffusion was 81% and 95.2%, and the specificity was 92.3% and 84.6%, respectively.
The blaVIM, blaKPC and blaNDM genes were detected in 18 (52.9%), 1 (2.9%) and 1 (2.9%) of the carbapenem-resistant P. aeruginosa isolates, respectively. Besides, 1 isolate (2.9%) carries two carbapenemase genes VIM+KPC. None of the isolates carry the bla OXA48 gene.
Virulence Factors
The frequency of evaluated virulence factors protease, pyocyanin, twitching motility, hemolytic activity and biofilm formation was 76.2%, 58.8%, 83.8%, 93.8% and 77.5%, respectively. All isolates of P. aeruginosa carry quorum-sensing lasR and rhlR genes. Biofilm formation was significantly associated with carbapenem resistance and MDR and XDR P. aeruginosa, while pyocyanin production was significantly correlated to carbapenem sensitive isolates. Antimicrobial resistance and virulence factors in carbapenem-resistant P. aeruginosa and the carbapenem-susceptible P. aeruginosa are presented in Table 2.
 |
Table 2 Comparison Of Antibiotic Resistance And Virulence Factors In Carbapenem-Resistant P. aeruginosa And The Carbapenem-Susceptible P. aeruginosa |
Virulence factors produced in MDR and non-MDR strains P. aeruginosa are illustrated in Figure 1.
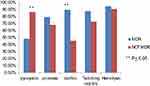 |
Figure 1 Virulence factors in MDR and non-MDR P. aeruginosa strains. |
Discussion
Multidrug-resistant P. aeruginosa is a rising health problem that limits the options for treatment and prolongs hospitalization.21 In our study, MDR and XDR P. aeruginosa represents 72.5% of isolates of P. aeruginosa. Furthermore, more than half of these MDR and XDR isolates were carbapenem resistant. A similar rate was reported in Venezuela in which MDR and XDR P. aeruginosa increased to 71.9% in 2016.22 Also, Rossi Goncalves23 concluded a high rate of MDR (73.9%). However, a lower rate of MDR was reported in other studies conducted in Egypt.24,25 This high rate of resistance among our isolates may be due to the fact that they were from hospital-acquired infections.26
The rate of imipenem resistance varies widely from 1% to 73.5%.12,16,23–25,27,28 In our study, the frequency of imipenem resistance in P. aeruginosa was 42.5%. Moreover, these isolates were significantly associated with resistance to other antibiotics such as cefepime, piperacillin-tazobactam and gentamicin. The same finding was previously reported.23,29 This high rate of imipenem resistance may be due to uncontrolled carbapenem use in hospital infections.22 Carbapenem resistance among P. aeruginosa isolates is attributed mainly to carbapenemase production.5 In the current study, 65% of carbapenem-resistant isolates carry carbapenemase genes, with blaVIM being the most common gene. Numerous studies reported that blaVIM gene is the most frequent MBL found in carbapenem-resistant P. aeruginosa.3,30 However, blaIMP gene was the most common detected carbapenemase gene in P. aeruginosa in a study conducted in Iran.29 Colistin and Polymyxin B showed a high rate of susceptibility and remain successful options for treatment of infections caused by MBL-producing P. aeruginosa.31 All our isolates were sensitive to colistin and Polymyxin B. In the absence of MBL enzymes, carbapenem resistance may be attributed to increasing production of AmpC chromosome-encoded cephalosporinase, increasing expression of efflux pump permeability and loss of porins.32
Virulence genes coexisting with resistance mechanisms to multiple antimicrobial in carbapenem-resistant P. aeruginosa have developed an emerging threat.3 On the other hand, other investigators found no certain associations between antibiotic resistance and virulence genes in MDR P. aeruginosa.30 Bogiel33 suggested that reduction of virulence in MDR P. aeruginosa may be owing to the fact that some genes selectively silence and activate other ones or reduced virulence of MDR strains is the suitable bacterial genome controlling that tolerates for existence in the presence of the antibiotic.
In this study, biofilm formation in carbapenem sensitive and carbapenem-resistant was 65.2% and 94.1%, respectively. Also, Ghanbarzadeh Corehtash34 demonstrated that the production of biofilm in MDR P. aeruginosa isolates was significantly higher than that in non–MDR P. aeruginosa isolates. Additionally, many authors described a significant positive correlation between MBL and AmpC β-lactamase production and biofilm formation.35 In contrast to our results, other studies found no significant difference in the association of biofilm formation and the presence of multidrug resistance.22,36 Also, no statistical significance was found between MBL and biofilm formation.31
Hemolysin and proteases were important virulence factors that were observed in 93.8% and 76.2% of our isolates. No significant difference was detected between carbapenem-resistant and carbapenem-sensitive P. aeruginosa for either production of hemolysin or protease or twitching motility. The same finding was formerly reported.25,37,38 Pyocyanin is a characteristic pigment produced by P. aeruginosa.19 In our isolates, pyocyanin production was significantly reduced in both carbapenem-resistant and MDR isolates. Likewise, other authors revealed that pyocyanin production by P. aeruginosa is reduced in MDR strains; furthermore, transduction of MBL genes into non-MDR P. aeruginosa strain production of pyocyanin reduced.36,39 But, others found that imipenem resistance was associated with a lack of alkaline protease production but not associated with a reduction in pyocyanin production.40
Several virulence factors investigated in this work such as alkaline protease, pyocyanin and biofilm formation were controlled by (QS) systems.40 A previous study found that there is an association between QS deficiency and decreased susceptibility to antimicrobials.40 In our study, all isolates of P. aeruginosa carry lasR and rhl genes. Likewise, QS genes were detected in all isolates from wound and respiratory tract secretions from infected patients who underwent cardiovascular surgery.7 QS deficient strains that fail to create successful infection were related to a decrease in the making of virulence factors.41 A major limitation of the study is that the gene expression of quorum sensing was not assessed.
Conclusion
Carbapenem resistance in P. aeruginosa represents a problem in hospital-acquired infections. The magnitude of this problem may arise if associated with other antibiotic resistance that limits options for treatment or presence of virulence factors that may increase the severity of the infection. Carbapenem-resistant P. aeruginosa was significantly associated with other antimicrobial resistance and biofilm formation but shows reduced production of pyocyanin. Surveillance program is needed to trace the origin and limit transport to other patients.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Xavier DE, Picão RC, Girardello R, Fehlberg LC, Gales AC. Efflux pumps expression and its association with porin down-regulation and β-lactamase production among Pseudomonas aeruginosa causing bloodstream infections in Brazil. BMC Microbiol. 2010;10(1):217.
2. Poole K. Pseudomonas aeruginosa: resistance to the max. Front Microbiol. 2011;2:65. doi:10.3389/fmicb.2011.00065
3. Ellappan K, Belgode Narasimha H, Kumar S. Coexistence of multidrug resistance mechanisms and virulence genes in carbapenem-resistant Pseudomonas aeruginosa strains from a tertiary care hospital in South India. J Glob Antimicrob Resist. 2018;12:37–43. doi:10.1016/j.jgar.2017.08.018
4. Kim H, Sung JY, Yong D, et al. Disk carbapenemase test for the rapid detection of KPC-, NDM-, and other metallo-beta-lactamase-producing gram-negative bacilli. Ann Lab Med. 2016;36(5):434–440. doi:10.3343/alm.2016.36.5.434
5. Pollini S, Fiscarelli E, Mugnaioli C, et al. Pseudomonas aeruginosa infection in cystic fibrosis caused by an epidemic metallo-β-lactamase-producing clone with a heterogeneous carbapenem resistance phenotype. Clin Microbiol Infect. 2011;17(8):1272–1275. doi:10.1111/j.1469-0691.2011.03466.x
6. Murray PR, Rosenthal KS, Pfaller MA. Medical Microbiology.
7. Cotar AI, Chifiriuc MC, Banu O, Lazar V. Molecular characterization of virulence patterns in Pseudomonas aeruginosa strains isolated from respiratory and wound samples. Biointerface Res Appl Chem. 2013;3(2).
8. Persyn E, Sassi M, Aubry M, et al. Rapid genetic and phenotypic changes in Pseudomonas aeruginosa clinical strains during ventilator-associated pneumonia. Sci Rep. 2019;9(1):4720. doi:10.1038/s41598-019-41201-5
9. Mahon C, Lehman D, Manuselis GJMEHS
10. Clinical laboratory Institute standard. Document M100-S25 Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fifth Informational Supplement. Wayne: Clinical and Laboratory Standards Institute; 2015.
11. Clinical laboratory Institute standard. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically; Approved Standard—9th Edn (CLSI Document M07–A9). Wayne, PA: Clinical and Laboratory Standards Institute; 2012.
12. Magiorakos AP, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–281. doi:10.1111/j.1469-0691.2011.03570.x
13. Lee K, Chong Y, Shin HB, Kim YA, Yong D, Yum JH. Modified Hodge and EDTA-disk synergy tests to screen metallo-beta-lactamase-producing strains of Pseudomonas and Acinetobacter species. Clin Microbiol Infect. 2001;7(2):88–91. doi:10.1046/j.1469-0691.2001.00204.x
14. Yong D, Lee K, Yum JH, Shin HB, Rossolini GM, Chong Y. Imipenem-EDTA disk method for differentiation of metallo-beta-lactamase-producing clinical isolates of Pseudomonas spp. and Acinetobacter spp. J Clin Microbiol. 2002;40(10):3798–3801. doi:10.1128/JCM.40.10.3798-3801.2002
15. Poirel L, Walsh TR, Cuvillier V, Nordmann P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. 2011;70(1):119–123. doi:10.1016/j.diagmicrobio.2010.12.002
16. Zhu H, Bandara R, Conibear TC, et al. Pseudomonas aeruginosa with lasI quorum-sensing deficiency during corneal infection. Invest Ophthalmol Vis Sci. 2004;45(6):1897–1903. doi:10.1167/iovs.03-0980
17. Castro-Escarpulli G, Figueras MJ, Aguilera-Arreola G, et al. Characterisation of Aeromonas spp. isolated from frozen fish intended for human consumption in Mexico. Int J Food Microbiol. 2003;84(1):41–49. doi:10.1016/S0168-1605(02)00393-8
18. Kus JV, Tullis E, Cvitkovitch DG, Burrows LLJM. Significant differences in type IV pilin allele distribution among Pseudomonas aeruginosa isolates from cystic fibrosis (CF) versus non-CF patients. Microbiology. 2004;150(5):1315–1326. doi:10.1099/mic.0.26822-0
19. Essar DW, Eberly L, Hadero A, Crawford I. Identification and characterization of genes for a second anthranilate synthase in Pseudomonas aeruginosa: interchangeability of the two anthranilate synthases and evolutionary implications. J Bacteriol. 1990;172(2):884–900. doi:10.1128/jb.172.2.884-900.1990
20. O’Toole GAJJ. Microtiter dish biofilm formation assay. J Vis Exp. 2011;47:e2437.
21. Gaynes R, Edwards JR. Overview of nosocomial infections caused by gram-negative bacilli. Clin Infect Dis. 2005;41(6):848–854. doi:10.1086/432803
22. Rodulfo H, Arcia A, Hernandez A, et al. Virulence factors and integrons are associated with MDR and XDR phenotypes in nosocomial strains of Pseudomonas aeruginosa in a Venezuelan university hospital. Rev Inst Med Trop Sao Paulo. 2019;61:e20. doi:10.1590/s1678-9946201961020
23. Rossi Goncalves I, Dantas RCC, Ferreira ML, Batistao D, Gontijo-Filho PP, Ribas RM. Carbapenem-resistant Pseudomonas aeruginosa: association with virulence genes and biofilm formation. Braz J Microbiol. 2017;48(2):211–217. doi:10.1016/j.bjm.2016.11.004
24. Hashem H, Hanora A, Abdalla S, Shaeky A, Saad A. Dissemination of metallo-beta-lactamase in Pseudomonas aeruginosa isolates in Egypt: mutation in blaVIM-4. APMIS. 2017;125(5):499–505. doi:10.1111/apm.12669
25. Sonbol FI, Khalil MAEF, Mohamed AB, Ali SS. Correlation between antibiotic resistance and virulence of pseudomonas aeruginosa clinical isolates. Turk J Med Sci. 2015;45:568–577. doi:10.3906/sag-1406-58
26. Strateva T, Yordanov D. Pseudomonas aeruginosa - a phenomenon of bacterial resistance. J Med Microbiol. 2009;58(Pt 9):1133–1148. doi:10.1099/jmm.0.009142-0
27. Ruiz-Roldan L, Belles A, Bueno J, et al. Pseudomonas aeruginosa isolates from Spanish children: occurrence in faecal samples, antimicrobial resistance, virulence, and molecular typing. Biomed Res Int. 2018;2018:8060178. doi:10.1155/2018/8060178
28. Silva LV, Galdino AC, Nunes AP, et al. Virulence attributes in Brazilian clinical isolates of Pseudomonas aeruginosa. Int J Med Microbiol. 2014;304(8):990–1000. doi:10.1016/j.ijmm.2014.07.001
29. Dogonchi AA, Ghaemi EA, Ardebili A, Yazdansetad S, Pournajaf A. Metallo-beta-lactamase-mediated resistance among clinical carbapenem-resistant Pseudomonas aeruginosa isolates in northern Iran: a potential threat to clinical therapeutics. Ci Ji Yi Xue Za Zhi. 2018;30(2):90–96. doi:10.4103/tcmj.tcmj_101_17
30. Al Dawodeyah HY, Obeidat N, Abu-Qatouseh LF, Shehabi AA. Antimicrobial resistance and putative virulence genes of Pseudomonas aeruginosa isolates from patients with respiratory tract infection. Germs. 2018;8(1):31–40. doi:10.18683/germs.2018.1130
31. Baniya B, Pant ND, Neupane S, et al. Biofilm and metallo beta-lactamase production among the strains of Pseudomonas aeruginosa and Acinetobacter spp. at a Tertiary Care Hospital in Kathmandu, Nepal. Ann Clin Microbiol Antimicrob. 2017;16(1):70. doi:10.1186/s12941-017-0245-6
32. Livermore DM. Of Pseudomonas, porins, pumps and carbapenems. J Antimicrob Chemother. 2001;47(3):247–250. doi:10.1093/jac/47.3.247
33. Bogiel T, DEPTUŁA A, KWIECIŃSKA-PIRÓG J, PRAŻYŃSKA M, Mikucka A, Gospodarek-Komkowska E. The prevalence of exoenzyme S gene in multidrug-sensitive and multidrug-resistant Pseudomonas aeruginosa clinical strains. Pol J Microbiol. 2017;66(4):427–431. doi:10.5604/01.3001.0010.6500
34. Ghanbarzadeh Corehtash Z, Khorshidi A, Firoozeh F, Akbari H, Mahmoudi Aznaveh A. Biofilm formation and virulence factors among pseudomonas aeruginosa isolated from burn patients. Jundishapur J Microbiol. 2015;8(10):e22345. doi:10.5812/jjm
35. Heydari S, Eftekhar F. Biofilm formation and β-lactamase production in burn isolates of Pseudomonas aeruginosa. Jundishapur J Microbiol. 2015;8(3):e15514–e15514. doi:10.5812/jjm
36. Deptula A, Gospodarek E. Reduced expression of virulence factors in multidrug-resistant Pseudomonas aeruginosa strains. Arch Microbiol. 2010;192(1):79–84. doi:10.1007/s00203-009-0528-1
37. Jacome PR, Alves LR, Cabral AB, Lopes AC, Maciel MA. Phenotypic and molecular characterization of antimicrobial resistance and virulence factors in Pseudomonas aeruginosa clinical isolates from Recife, State of Pernambuco, Brazil. Rev Soc Bras Med Trop. 2012;45(6):707–712. doi:10.1590/S0037-86822012000600010
38. Wang H, Tu F, Gui Z, Lu X, Chu W. Antibiotic resistance profiles and quorum sensing-dependent virulence factors in clinical isolates of pseudomonas aeruginosa. Indian J Microbiol. 2013;53(2):163–167. doi:10.1007/s12088-013-0370-7
39. Fuse K, Fujimura S, Kikuchi T, et al. Reduction of virulence factor pyocyanin production in multidrug-resistant Pseudomonas aeruginosa. J Infect Chemother. 2013;19(1):82–88. doi:10.1007/s10156-012-0457-9
40. Karatuna O, Yagci A. Analysis of quorum sensing-dependent virulence factor production and its relationship with antimicrobial susceptibility in Pseudomonas aeruginosa respiratory isolates. Clin Microbiol Infect. 2010;16(12):1770–1775. doi:10.1111/j.1469-0691.2010.03177.x
41. Kumar R, Chhibber S, Harjai K. Quorum sensing is necessary for the virulence of Pseudomonas aeruginosa during urinary tract infection. Kidney Int. 2009;76(3):286–292. doi:10.1038/ki.2009.183
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
