Back to Journals » Infection and Drug Resistance » Volume 12
Virulence and genomic features of a blaCTX-M-3 and blaCTX-M-14 coharboring hypermucoviscous Klebsiella pneumoniae of serotype K2 and ST65
Authors Fu Y, Xu M, Liu Y, Li A, Zhou J
Received 12 September 2018
Accepted for publication 5 November 2018
Published 3 January 2019 Volume 2019:12 Pages 145—159
DOI https://doi.org/10.2147/IDR.S187289
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Joachim Wink
Yiqi Fu,1,* Min Xu,2,* Yanchao Liu,2 Ang Li,3 Jianying Zhou1
1Department of Respiratory Diseases, The First Affiliated Hospital, College of Medicine, Zhejiang University, Hangzhou, China; 2Key Laboratory of Clinical In Vitro Diagnostic Techniques of Zhejiang Province, Center of Clinical Laboratory, The First Affiliated Hospital, College of Medicine, Zhejiang University, Hangzhou, China; 3Department of Precision Medicine Center, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, China
*These authors contributed equally to this work
Background: Capsular serotype K2 Klebsiella pneumoniae of sequence type (ST) 65 has been recognized as a hypervirulent clone. Simultaneous presence of different blaCTX-M genes has never been reported in this clone. In the present study, the genetic characteristics and virulence phenotype of a CTX-M-3 and CTX-M-14 coproducing ST65 K. pneumoniae human isolate, KP_06, that caused an intracranial infection, are evaluated.
Methods: The potential virulence of KP_06 was assayed by in vitro and in vivo methods. The molecular biology and whole-genome sequencing technology were used to analyze the genomic features associated with the virulence of this strain.
Results: The KP_06 exhibited typical features of hypervirulent K. pneumoniae (hvKP), showing hypermucoviscosity phenotype and belonging to K2 and ST65. Apart from virulence genes linked to hvKP, including rmpA, rmpA2, and clb cluster and genes encoding siderophores, it was found to harbor a ~170 kb pLVPK-like virulence plasmid. In contrast to most hvKP, KP_06 was resistant to cephalosporins and the coexistence of blaCTX-M-3 and blaCTX-M-14 was detected. Further experiments demonstrated that this strain was classified as a nonbiofilm producer and serum sensitivity (grade 1) and killed only 30% of Galleria mellonella inoculated with 1×106 colony-forming unit of the specimen within 48 hours, suggesting relatively low virulence. Comparative genomic analysis of KP_06 with five K2 hypermucoviscous K. pneumoniae (HMKP) revealed seven unique orthologies with varied function in this strain. Intriguingly, the virulence genes identified in KP-06 were unexpectedly more diverse than those observed in five other K2 HMKP strains.
Conclusion: Our data support the notion that neither virulence-associated genes (clusters) nor the pLVPK-like virulence plasmid is sufficient for the hypervirulence of K. pneumoniae. Future studies aiming to explore the virulence of K. pneumoniae should take genome-based profile together with experimental work. The detailed mechanism involving in the impaired virulence of KP_06 remains to be further explored.
Keywords: Klebsiella pneumoniae, virulence factor, serotype K2, ST65, blaCTX-M, comparative genome
Introduction
Since first identified in 1986 in Taiwan, hypervirulent Klebsiella pneumoniae (hvKP), which caused severe invasive infections such as liver abscesses, endophthalmitis, meningitis, osteomyelitis, and necrotizing fasciitis in otherwise healthy individuals,1 has been increasingly reported and become the focus of concern recent years.
Traditionally, a positive string test, which is defined as hypermucoviscosity (HM) phenotype appeared to be a surrogate marker for hvKP.2 However, new evidence has suggested that HM and hypervirulence are two distinct phenotypes of K. pneumoniae that should not be used synonymously.3 Most recently, new genetic biomarkers for hvKP such as iucA located on the virulence plasmid encoding for aerobactin have been explored.4 Hence, the precise definition of hvKP remains controversial and hypervirulence-associated determinants of K. pneumoniae required further study.
Isolates with serotypes K1 and K2 have been demonstrated as particularly virulent.5 Compared to serotype K1 strains mainly associated with sequence type (ST) 23, K. pneumoniae strains with serotype K2 exhibited more diverse genetic types, among which ST65, ST86, and ST380 are predominant and considered to be hypervirulent clones.6 The serotype K1 or K2 K. pneumoniae strains have not generally been associated with acquired antimicrobial resistance (AMR), but the last few years increasing reports of resistant strains, including those resistant to third-generation cephalosporins and even carbapenems, were observed.7–9 Up to now, the AMR genes including blaCTX-M-3,7 blaSHV-148,10 blaKPC-2,11 and blaNDM12 have been found in K2 serotype K. pneumoniae strains, whereas the simultaneous presence of different blaCTX-M genes in the hypermucoviscous K. pneumoniae (HMKP) of serotype K2 has never been reported.
As the application of whole-genome sequencing (WGS) in clinical microbiology extensively, the genomes of many human source K. pneumoniae have been sequenced. However, to the best of our knowledge, only five genomic sequences of serotype K2 K. pneumoniae with varied genetic backgrounds were publicly available up to now (RJF293, U25, 52.145, hvKP1, and NUHL24835).12–16 In the present study, we assayed the potential virulence and characterized the genomic features of a blaCTX-M-3 and blaCTX-M-14 coharboring K. pneumoniae strain of K2 isolated from human cerebrospinal fluid (CSF). We also compared the genome sequences of KP_06 with five other K2 K. pneumoniae strains aiming to investigate the strain-specific genes. Our finding will make up our current knowledge on the evolutionary relationship between virulence and resistance in serotype K2 K. pneumoniae.
Methods
Isolates and antimicrobial susceptibility testing
The VITEK-2 compact system (bioMérieux, Craponne, France) was used to establish the strain identification and antimicrobial susceptibility testing. The results were interpreted in accordance with the guideline document M100-S28 established by Clinical and Laboratory Standards Institute.17 The species identification of the isolate was then confirmed by matrix-assisted laser desorption/ionization mass spectrometry (Bruker Optics Inc, Billerica, MA, USA). Escherichia coli American Type Culture Collection (ATCC) 25922 and K. pneumoniae 700603 were used as quality control.
In the in vitro and in vivo virulence assessments, an HM ST23:K1 K. pneumoniae strain (KP_07), which isolated from pus of a patient with liver abscess with wild-type susceptibility profile to antibiotics, was used as the positive control. A KPC-2-producing ST11 K. pneumoniae strain, KP_36, without aerobactin-encoding genes, rmpA and rmpA2, was used as the control for low virulence as previously described.18
Detection of resistance mechanisms and virulence-associated factors
The ESBL genes (blaCTX-M and blaSHV) and carbapenemase genes (blaKPC, blaNDM, blaVIM, and blaOXA-48) were amplified by PCR. The amplicons of β-lactamase genes were purified and sequenced in an ABI 3730 DNA sequencer (Thermo Fisher Scientific, Waltham, MA, USA). The sequences obtained were compared with those in the NCBI database using the Blast software (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
The HM phenotype was determined by “string test”. Briefly, when using a bacteriology loop to stretch bacterial colony cultured on an agar plate overnight at 37°C, the generation of a viscous string of >5 mm in length was defined as positive.19 The presence of serotype-specific genes encoding for K1, K2, K5, K20, K54, and K57 and 14 known K. pneumoniae virulence genes, including magA, rmpA, rmpA2, kfu, allS, fimH, wabG, ybtS, mrkD, uge, entB, iutA, iucA, and iroN, were assessed as previously described.20,21
S1 nuclease pulsed-field gel electrophoresis (S1-PFGE) and southern blot hybridization
The number of plasmids and the size of the pLVPK-like virulence plasmid carried by K. pneumoniae strains were determined by S1-PFGE and southern blot hybridization. In brief, whole DNA of K. pneumoniae was subjected to S1 nuclease (Takara Bio Inc., Tokyo, Japan) digestion. Digested fragments were subjected to PFGE. Then, the gels were blotted onto nylon membranes (EMD Millipore, Billerica, MA, USA) according to standard techniques. The membranes were hybridized with digoxigenin-labeled rmpA2 probe.
Growth curve assays
Growth curve was generated by diluting equal numbers of colony-forming unit (CFU) of each isolate (approximately 1×106 CFU/mL) in Luria-Bertani (LB) broth and incubated in 96-well microplates. Then, the growth was measured by OD600 nm using a microplate reader (Synergy HT; Biotek Instruments Inc., Winooski, VT, USA) equipped with the Gen5 Microplate software (Biotek Instruments). Each curve was performed in triplicate.
Biofilm formation assays
Biofilm formation was assessed by the method of Araújo et al.22 A total of 1 µL of bacterial suspensions obtained from overnight culture was inoculated into 100 µL of fresh LB broth in individual wells of a 96-well flat-bottomed polystyrene plate. After 24 hours of static incubation at 37°C, the biomass was measured by crystal violet staining and acetic acid elution. The absorbance for the eluted dye was determined at OD570 nm using the microplate reader (Synergy HT). The results were interpreted according to the criteria established by Saxena et al.23 Each assay was performed at least three times at three occasions.
Susceptibility to serum killing
The bacterial susceptibility to serum was performed as described previously.12 Briefly, bacteria grown in LB broth were collected during the mid-log phase, then diluted to 1×106 CFU/mL. A total of 25 µL of the bacterial suspension were mixed with 75 µL of pooled healthy human serum in microtiter plate and then incubated in 37°C with gently shake. Serial dilutions were plated on Mueller-Hinton (MH) agar at 0, 1, 2, and 3 hours to obtain the colony counts of viable bacteria. The results were graded from 1 to 6 as described by Mei et al,12 with grade 1 considered to be the most serum susceptibility. Each strain was tested at least three times.
Infection of Galleria mellonella larvae
Bacteria from overnight cultures were washed with PBS and adjusted to concentrations 1×104, 1×105, 1×106, and 1×107 CFU/mL in PBS. Ten randomly selected Wax moth larvae (G. mellonella) weighing between 250 and 350 mg were for each isolate of each concentration. The insects were injected 10 µL of bacterial suspension via rear left proleg and then incubated in dark at 37°C. The numbers of dead insects were recorded every 12 hours for 48 hours. PBS injection controls and controls receiving no injection were used to evaluated trauma and attrition, respectively.
WGS and data analysis
The KP_06 strain was subjected to WGS. Genomic DNA was prepared using the QIAamp DNA Mini Kit (Qiagen NV, Venlo, the Netherlands), and the whole genomic sequencing was conducted using the Illumina HiSeq 2500-PE125 platform. The low-quality reads were filtered and then assembled into scaffolds using SOAP de novo. The ST of KP_06 was identified using the Center for Genomic Epidemiology (CGE) service (https://cge.cbs.dtu.dk/services/MLST/),24 and the serotype was determined by querying the wzi allele using Pasteur database (http://bigsdb.pasteur.fr/klebsiella/klebsiella.html). AMR genes were identified from the genome sequences against the Resfinder (https://cge.cbs.dtu.dk/services/ResFinder/)25 and the Comprehensive Antibiotic Resistance Database (CARD) Version 2.0.1 (https://card.mcmaster.ca/).26 The putative virulence factors were predicted by the Virulence Factor Database (VFDB) protein sequences of core data set (Version April 28, 2018, 2,595 genes, http://www.mgc.ac.cn/cgi-bin/VFs/v5/main.cgi)27 and virulence genes deposited in K. pneumoniae multi-locus sequence typing (MLST) database of Institut Pasteur (http://bigsdb.pasteur.fr/klebsiella/klebsiella.html) with an e-value cutoff of e−10 and an identify threshold of 60%. For comparative genomic analysis, the five available genome sequences of K2 K. pneumoniae (RJF293, U25, 52.145, hvKP1, and NUHL24835) were retrieved from the GenBank and the pairwise single-nucleotide polymorphisms’ (SNPs) analysis was carried out using the CSI Phylogeny 1.4 pipeline available at https://cge.cbs.dtu.dk/services/CSIPhylogeny/. Multiple genome alignment was performed using Mauve and BRIG programs. The Shared orthologies between the six K2 K. pneumoniae strains were identified by custom python script then illustrated in network graph by using Cytoscape Version 3.5.1. Finally, the function of ortholog clusters was classified according to clusters of orthologous groups (COGs).
This Whole-Genome Shotgun project has been deposited at GenBank under the accession number QBUJ00000000. The accession numbers for the genomes of strains RJF293, U25, 52.145, hvKP1, and NUHL24835 are CP014008, CP012043, FO834906, AOIZ00000000, and CP014004, respectively.
Ethics approval and informed consent
This study was conducted in accordance with the Declaration of Helsinki, and the ethical approval was granted from the Ethics Committees and Review Board of the First Affiliated Hospital, College of Medicine, Zhejiang University. Written informed consent of this case was obtained from the patient for publication.
Results
Patient characteristics
A 66-year-old woman was admitted to our hospital with numbness in her left lower limb for 3 days. She was accompanied by type 2 diabetes mellitus for 5 years and had a history of hypertension for 18 years. After MRI of the cervical spine and skull base, significant herniation of the cerebellar tonsils was observed, which calling for surgery intervention. Ceftriaxone 2.0 g once was given intravenous before the surgery for prophylaxis, and vancomycin 0.5 g Q8H was given after surgery for prophylaxis. The first day after surgery, the patient developed occipital headache and fever. The CSF examination is positive, with cloudy appearance, substantially increased polymorphonuclear (PMN) cells (3,200/mm3; 92% segmented neutrophils and 8% lymphocytes) and significantly elevated protein level (2.66 g/L). A presume diagnosis of meningitis was made. Empiric treatment of intravenous imipenem (0.5 g Q8H) and vancomycin (0.5 g Q6H) was initiated immediately. Culture of the CSF grew K. pneumoniae, designated as KP_06. Two days later, the temperature elevation resolved and repeated lumbar puncture yielded slight cloudy CSF with improved parameters: PMN 180/mm3 and protein 1.2 g/L. The CSF culture remained negative for K. pneumoniae, and thereafter, until the patient was discharged 73 days later.
Microbiological characteristics of KP_06
The KP_06 strain showed resistance to cephalosporins (ceftriaxone, ceftazidime, and cefepime), β-lactam/β-lactamase inhibitor combination (ampicillin–sulbactam and piperacillin–tazobactam) and aztreonam, while susceptible to fluoroquinolones, carbapenems, and tigecycline (Table S1). The coexistence of blaCTX-M-3 and blaCTX-M-14 in this strain was subsequently identified by PCR and sequencing.
A viscous string longer than 30 mm by the string test was observed in the KP_06 strain, suggesting HM phenotype. The strain was determined to be ST65 (gapA–infB–mdh–pgi–phoE–rpoB–tonB: 2–1–2–1–10–4–13) and capsular serotype K2 by PCR and sequencing. Several virulence genes including fimH, wabG, ybtS, mrkD, uge, and entB and the pLVPK-derived loci (rmpA, rmpA2, iucA, iutA, and iroN) were detected in KP_06. A pLVPK-like virulence plasmid of approximately 170 kb was further confirmed by hybridization of S1-digested DNA with rmpA2 probe (Figure S1).
In vitro and in vivo virulence assessment of KP_06
The growth rates between KP_06 and other two strains (KP_07: ST23:K1 strain and KP_36: KPC-2-producing ST11 K. pneumoniae strain) were not differently significant (Figure S2).
Figure 1A depicts the ability of biofilm formation of the strains. The level of biofilm formation of KP_06 was similar to that of KP_36, while significantly lower than that of KP_07 (OD570 nm, 0.29±0.01 vs 0.83±0.10, P<0.001). According to the criteria established by Saxena et al,23 the cutoff OD value (ODC) in our study was determined as OD570 nm = 0.29, hence the KP_06 and KP_36 were identified as nonbiofilm producers (OD570 nm ≤ 0.29), whereas the KP_07 was moderate producer (0.58 ≤ OD570 nm = 0.83≤1.16).
Serum killing sensitive grades 1 and 2 were found in KP_06 and KP_36 strains, respectively, whereas KP_07 showed serum resistance (grade 6) (Figure 1B). Consistent with the results of biofilm formation and serum killing assays, the KP_06 strain produced very low mortality rate in G. mellonella, leading to about 70% survival of G. mellonella at 48 hours after being challenged with 106 CFU of bacteria. The KP_36 showed to be more virulent than KP_06, resulting in 50% survival at 48 hours after being challenged with 106 CFU of bacteria. The classic ST23:K1 strain KP_07 resulted in 0% survival at 36 hours, displaying the most virulent strain (Figure 1C). Data about the effects of other inoculums of the strains are available in Figure S3.
Comparative genomic analysis
The genome sequence of KP_06 was 5,655,975 bp in size and had a GC content of 56.9% in 65 contigs, with N50 spanning 259,669 bp. The general genomic features of KP_06 are listed in Table 1. Although KP_06 and five other publicly available K2 K. pneumoniae strains (RJF293, U25, 52.145, hvKP1, and NUHL24835) displayed HM phenotype, they belonged to varied STs (Table 2). Pairwise SNP analysis for these six strains based on their raw sequencing reads showed that their core genome differed much from each other (Table S2), which consistent with the results of comparative chromosome analysis visualized by the circular maps (Figure 2A). Further comparative genomic analysis identified 1,378 shared orthologies among the six K2 K. pneumoniae strains, whereas the KP_06 contained seven unique orthologies (Figure 2B). We also analyzed the functional classification of ortholog clusters using the COGs’ database. The results obtained are summarized in Table 3. The most abundant category in the shared orthologies was amino acid transport and metabolism (172), followed by carbohydrate transport and metabolism (140), and inorganic ion transport and metabolism (124). The unique orthologies of KP_06 were identified as involved in (V) defense mechanisms, (Z) cytoskeleton, (M) cell wall/membrane/envelope biogenesis, (H) coenzyme transport and metabolism, (D) cell cycle control, cell division, and chromosome partitioning, (I) lipid transport and metabolism, (G) carbohydrate transport and metabolism, and (S) function unknown.
  | Table 1 General genomic features of the KP_06 isolate Abbreviation: CO, clusters of orthologous groups. |
  | Table 3 The distribution of COGs’ functional catalogs of the orthologies for the six K. pneumoniae strains Abbreviations: COGs, clusters of orthologous groups; K. pneumoniae, Klebsiella pneumoniae. |
Detection of virulence and antibiotic-resistant genes profiles by WGS
Multiple genes have been presumed associated with increased virulence in K. pneumoniae strains, hence WGS was used to identify virulence genes previously described as linked to hvKP against publicly available databases. All the six K2 strains carried the type 1 and type 3 fimbrial gene clusters (mrk and fim), the genes encoding for yersiniabactin (ybt and irp complex) and the genes involved in the synthesis of enterobactin (ent and fep). The detailed virulence genes detected are listed in Table S3. Although in vitro and in vivo virulence assays showed that the virulence of KP_06 was low, this strain unexpectedly carried more abundant of putative virulence genes (118 virulence genes) compared with five other K2 strains. It also harbored genes including iucABCD-iutA, iroBCDN, rmpA, and rmpA2, which usually located in the pLVPK-like virulence plasmid. This observation was consistent with the results of hybridization analysis, wherein the rmpA2-encoding virulence plasmid was present in KP_06 and KP_07 but not in the KP_36 strain (Figure S1). Additionally, the colibactin synthesis locus clb was detected in KP_06 and RJF293.
Although it is common sense that AMR genes were rarely found in HMKP strains, β-lactam resistance genes were identified in all of the analyzed K2 strains except KP52.145. Besides the simultaneous presence of different blaCTX-M (blaCTX-M-3 and blaCTX-M-14) in KP_06, the presence of other β-lactam-resistant genes blaSHV-11 and blaTEM-1, fluoroquinolone-resistant gene qnrS1, and fosfomycin-resistant gene fosA was also detected (Table 2).
Discussion
During the past decades, K. pneumoniae especially hvKP was recognized as the leading causative pathogen for adult bacterial meningitis in several Asian regions such as Taiwan28 and South Korea.29 However, to the best of our knowledge, there was only one case reporting suspected hvKP meningitis in mainland China up to now and the detailed virulence and genomic features of the causing pathogen were not investigated.30 In the present study, the phenotypic and genomic features of a K2 HMKP causing meningitis were characterized.
The patient developing postneurosurgical meningitis was accompanied by diabetes mellitus and hypertension, both of which were well-documented predisposing factors for hvKP infections.31,32 The KP_06 strain subsequently isolated from CSF exhibited HM nature and carried critical virulence determinants such as HM phenotype regulators (rmpA/rmpA2) and siderophore encoding systems (aerobactin, enterobactin, salmochelin, and yersiniabactin). The association between mortality and mucoid phenotype in K2 K. pneumoniae strain had been established in infected mouse model by Yu et al.5 Thereafter, the terms of HMKP and hvKP have often been used synonymously.3 As the in-depth research of virulence mechanisms of this new variant, siderophores especially aerobactin are considered integral to bacterial virulence due to its ability allowing bacteria to scavenge for iron from host transport proteins.33 Hence, recent studies viewed genetic factors associated with HM together with iron acquisition as unequivocal marker for hvKP identification.34,35 However, in the current case, even harboring these typical features of hvKP, the KP_06 was assigned to a lowly virulent strain by comprehensive virulence assays. Therefore, future studies focus on hvKP calling for a combination of phenotype, genomic data, and experimental virulence. The identification of biomarkers with high accuracy for hvKP would be very imperative.
K. pneumoniae isolates with high degrees of virulence and drug resistance have emerged.7,8,11,12 Our analysis showed that ESBL genes were detected in four K2 strains (KP_06, RJF293, U25, and NUHL24835), which indicated that the presence of AMR genes in serotype K2 K. pneumoniae might be underestimated previously. The successful transformation of plasmids carrying blaCMY-2, blaKPC-2, or blaDHA-1 genes into hvKP has been documented.8,36,37 Otherwise, a carbapenem-resistant K. pneumoniae strain that acquired part of an hvKP virulence plasmid causing a lethal nosocomial outbreak has been reported recently.21 Given the KP_06 belonged to the K2 and ST65 hypervirulent clone, we speculated it might be hvKP that acquired plasmids harboring different blaCTX-M genes.
As descripted by several studies,18 we chose a wild-susceptible hypermucoviscous ST23:K1 K. pneumoniae strain KP_07 as the control of hypervirulence in virulence assays. The KP_06 strain displayed much weaker capability of biofilm formation, serum resistance, and lethality compared to KP_07. Lavigne et al38 demonstrated that the presence of blaKPC was accompanied by impaired virulence of the isolates in Caenorhabditis elegans model. Choi et al39 also illustrated that the acquisition of colistin resistance leaded to the loss of HM phenotype, resulting in a decreased virulence in hvKP. These findings suggested that acquiring antibiotic resistance might impair the virulence of the clinical strains. However, comparative genomic analysis showed the virulence genes were more abundance in KP_06 than in other K2 hvKP (RJF293, 52.145, and NUHL24835). In addition, the presence of pLVPK-like virulence plasmid in this strain was confirmed. These results implied that neither possessing a virulence plasmid nor having virulence-associated genes (clusters) is equal to a higher level of virulence. Recently, Palacios et al40 reported the mutation of two marR (multiple antibiotic resistance regulator) homologous genes, kvrA and kvrB, significantly impaired the virulence of K1 and K2 serotype hvKP strains through reducing the expression of capsule synthesis associated genes. Moreover, a strong correlation between the quantitative siderophore production and in vivo virulence of K. pneumoniae has also been identified by Russo et al.4 Hence, it was reasonable to speculate that the decreased virulence of KP_06 might attribute to the low expression level of some critical virulence-associated determinants, but the exact mechanisms remain further exploration.
Although the in vitro and in vivo assays showed a decreased virulence of KP_06, the isolate caused a serious intracranial infection in a postoperative patient. One explanation was the disruption of skin and mucous barriers due to cervical spine surgery facilitated the invasion of bacteria, even if relatively low virulence isolate. Additionally, the actual virulence of bacteria in patients can be affected by the host environment at the site of infection. For instance, high glucose concentrations enhanced capsular polysaccharide biosynthesis of K. pneumoniae,41 which may happen in patients with diabetes mellitus. The biofilm formation significantly induced by the subinhibitory concentration of imipenem in Acinetobacter baumannii has also been reported.42 Further experiments to evaluate the virulence of KP_06 under stress conditions, such as antibiotics, are needed.
Conclusion
We investigated the virulence and genetic features of an HM K. pneumoniae strain (KP_06) collected from human CSF. Although displaying typical hypervirulent characteristics, this coharboring blaCTX-M-3 and blaCTX-M-14 K2 and ST65 K. pneumoniae was demonstrated to be a lowly virulent strain by in vitro and in vivo methods. However, the detailed mechanisms concerned with an impaired virulence of this strain remained further exploration. Although it is thought that the prevalence of K. pneumoniae strain with high degree of drug resistance and virulence will pose great threat to public health, our study still makes a valuable addition to the growing knowledge of the evolutionary relationship between resistance and virulence in K. pneumoniae.
Consent for publication
Written informed consent for publication of the individual details was obtained from the patient.
Acknowledgments
We thank the entire staff at the Department of Microbiology, The First Affiliated Hospital, College of Medicine, Zhejiang University, for their daily contributions to this study. We also thank the team of curators from the Institute Pasteur MLST (Paris, France) for importing novel alleles and profiles at http://bigsdb.pasteur.fr/klebsiella/klebsiella.html. This work was supported by the National Natural Science Foundation (no 81301459), Natural Science Foundation of Zhejiang Province (no LQ13H190001), Medical Science and Technology Project of Zhejiang Province (no 2014KYB096), and National Key Program for Infectious Diseases of China (no 2017ZX10103008).
Disclosure
The authors report no conflicts of interest in this work.
References
Lee CR, Lee JH, Park KS, et al. Antimicrobial resistance of hypervirulent Klebsiella pneumoniae: epidemiology, hypervirulence-associated determinants, and resistance mechanisms. Front Cell Infect Microbiol. 2017;7:483. | ||
Li W, Sun G, Yu Y, et al. Increasing occurrence of antimicrobial-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae isolates in China. Clin Infect Dis. 2014;58(2):225–232. | ||
Catalán-Nájera JC, Garza-Ramos U, Barrios-Camacho H. Hypervirulence and hypermucoviscosity: two different but complementary Klebsiella spp. phenotypes? Virulence. 2017;8(7):1111–1123. | ||
Russo TA, Olson R, Fang CT, et al. Identification of biomarkers for differentiation of hypervirulent klebsiella pneumoniae from classical K. pneumoniae. J Clin Microbiol. 2018;56(9):e00776-18. | ||
Yu VL, Hansen DS, Ko WC, et al. Virulence characteristics of Klebsiella and clinical manifestations of K. pneumoniae bloodstream infections. Emerg Infect Dis. 2007;13(7):986–993. | ||
Bialek-Davenet S, Criscuolo A, Ailloud F, et al. Genomic definition of hypervirulent and multidrug-resistant Klebsiella pneumoniae clonal groups. Emerg Infect Dis. 2014;20(11):1812–1820. | ||
Surgers L, Boyd A, Girard PM, Arlet G, Decré D. ESBL-producing strain of hypervirulent klebsiella pneumoniae K2, France. Emerg Infect Dis. 2016;22(9):1687–1688. | ||
Xie Y, Tian L, Li G, et al. Emergence of the third-generation cephalosporin-resistant hypervirulent Klebsiella pneumoniae due to the acquisition of a self-transferable blaDHA-1-carrying plasmid by an ST23 strain. Virulence. 2018;9(1):838–844. | ||
Zhang R, Lin D, Chan EW, Gu D, Chen GX, Chen S. Emergence of carbapenem-resistant serotype K1 hypervirulent Klebsiella pneumoniae strains in China. Antimicrob Agents Chemother. 2016;60(1):709–711. | ||
Zhang Y, Zhao C, Wang Q, et al. High prevalence of hypervirulent Klebsiella pneumoniae infection in China: geographic distribution, clinical characteristics, and antimicrobial resistance. Antimicrob Agents Chemother. 2016;60(10):6115–6120. | ||
Yao B, Xiao X, Wang F, Zhou L, Zhang X, Zhang J. Clinical and molecular characteristics of multi-clone carbapenem-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae isolates in a tertiary hospital in Beijing, China. Int J Infect Dis. 2015;37:107–112. | ||
Mei YF, Liu PP, Wan LG, et al. Virulence and genomic feature of a virulent Klebsiella pneumoniae sequence type 14 strain of serotype K2 harboring blaNDM-5 in China. Front Microbiol. 2017;8:335. | ||
Lery LM, Frangeul L, Tomas A, et al. Comparative analysis of Klebsiella pneumoniae genomes identifies a phospholipase D family protein as a novel virulence factor. BMC Biol. 2014;12:41. | ||
Rafiq Z, Sam N, Vaidyanathan R. Whole genome sequence of Klebsiella pneumoniae U25, a hypermucoviscous, multidrug resistant, biofilm producing isolate from India. Mem Inst Oswaldo Cruz. 2016;111(2):144–146. | ||
Wang X, Xie Y, Li G, et al. Whole-Genome-Sequencing characterization of bloodstream infection-causing hypervirulent Klebsiella pneumoniae of capsular serotype K2 and ST374. Virulence. 2018;9(1):510–521. | ||
Russo TA, Gill SR. Draft genome sequence of the hypervirulent klebsiella pneumoniae strain hvKP1, isolated in Buffalo, New York. Genome Announc. 2013;1(2):e0006513. | ||
CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 28th Informational Supplement (M100-S28). Wayne, PA: Clinical and Laboratory Standards Institute; 2018. | ||
Feng Y, Lu Y, Yao Z, Zong Z. Carbapenem-resistant hypervirulent Klebsiella pneumoniae of sequence type 36. Antimicrob Agents Chemother. 2018;62(7):e02644-17. | ||
Shon AS, Bajwa RP, Russo TA. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence. 2013;4(2):107–118. | ||
Compain F, Babosan A, Brisse S, et al. Multiplex PCR for detection of seven virulence factors and K1/K2 capsular serotypes of Klebsiella pneumoniae. J Clin Microbiol. 2014;52(12):4377–4380. | ||
Gu D, Dong N, Zheng Z, et al. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis. 2018;18(1):37–46. | ||
Araújo BF, Ferreira ML, Campos PA, et al. Hypervirulence and biofilm production in KPC-2-producing Klebsiella pneumoniae CG258 isolated in Brazil. J Med Microbiol. 2018;67(4):523–528. | ||
Saxena S, Banerjee G, Garg R, Singh M. Comparative study of biofilm formation in Pseudomonas aeruginosa isolates from patients of lower respiratory tract infection. J Clin Diagn Res. 2014;8(5):DC09– DC11. | ||
Larsen MV, Cosentino S, Rasmussen S, et al. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol. 2012;50(4):1355–1361. | ||
Zankari E, Hasman H, Cosentino S, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67(11):2640–2644. | ||
Jia B, Raphenya AR, Alcock B, et al. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017;45(D1):D566–D573. | ||
Chen L, Zheng D, Liu B, Yang J, Jin Q. VFDB 2016: hierarchical and refined dataset for big data analysis – 10 years on. Nucleic Acids Res. 2016;44(D1):D694–D697. | ||
Lee PY, Chang WN, Lu CH, et al. Clinical features and in vitro antimicrobial susceptibilities of community-acquired Klebsiella pneumoniae meningitis in Taiwan. J Antimicrob Chemother. 2003;51(4):957–962. | ||
Moon SY, Chung DR, Kim SW, et al. Changing etiology of community-acquired bacterial meningitis in adults: a nationwide multicenter study in Korea. Eur J Clin Microbiol Infect Dis. 2010;29(7):793–800. | ||
Qian Y, Wong CC, Lai SC, et al. Klebsiella pneumoniae invasive liver abscess syndrome with purulent meningitis and septic shock: a case from mainland China. World J Gastroenterol. 2016;22(9):2861–2866. | ||
Fang CT, Lai SY, Yi WC, Hsueh PR, Liu KL, Chang SC. Klebsiella pneumoniae genotype K1: an emerging pathogen that causes septic ocular or central nervous system complications from pyogenic liver abscess. Clin Infect Dis. 2007;45(3):284–293. | ||
Guo Y, Wang S, Zhan L, et al. Microbiological and clinical characteristics of hypermucoviscous Klebsiella pneumoniae isolates associated with invasive infections in China. Front Cell Infect Microbiol. 2017;7:24. | ||
Russo TA, Shon AS, Beanan JM, et al. Hypervirulent K. pneumoniae secretes more and more active iron-acquisition molecules than “classical” K. pneumoniae thereby enhancing its virulence. PLoS One. 2011;6(10):e26734. | ||
Li J, Ren J, Wang W, et al. Risk factors and clinical outcomes of hypervirulent Klebsiella pneumoniae induced bloodstream infections. Eur J Clin Microbiol Infect Dis. 2018;37(4):679–689. | ||
Yan Q, Zhou M, Zou M, Liu WE. Hypervirulent Klebsiella pneumoniae induced ventilator-associated pneumonia in mechanically ventilated patients in China. Eur J Clin Microbiol Infect Dis. 2016;35(3):3 87–396. | ||
Lin YT, Pan YJ, Lin TL, Fung CP, Wang JT. Transfer of CMY-2 cephalosporinase from escherichia coli to virulent klebsiella pneumoniae causing a recurrent liver abscess. Antimicrob Agents Chemother. 2015;59(8):5000–5002. | ||
Siu LK, Huang DB, Chiang T. Plasmid transferability of KPC into a virulent K2 serotype Klebsiella pneumoniae. BMC Infect Dis. 2014;14:176. | ||
Lavigne JP, Cuzon G, Combescure C, Bourg G, Sotto A, Nordmann P. Virulence of Klebsiella pneumoniae isolates harboring bla KPC-2 carbapenemase gene in a Caenorhabditis elegans model. PLoS One. 2013;8(7):e67847. | ||
Choi MJ, Ko KS, Ks K. Loss of hypermucoviscosity and increased fitness cost in colistin-resistant Klebsiella pneumoniae sequence type 23 strains. Antimicrob Agents Chemother. 2015;59(11):6763–6773. | ||
Palacios M, Miner TA, Frederick DR, et al. Identification of two regulators of virulence that are conserved in klebsiella pneumoniae classical and hypervirulent strains. MBio. 2018;9(4):e01443-18. | ||
Lee CH, Chen IL, Chuah SK, et al. Impact of glycemic control on capsular polysaccharide biosynthesis and opsonophagocytosis of Klebsiella pneumoniae: implications for invasive syndrome in patients with diabetes mellitus. Virulence. 2016;7(7):770–778. | ||
Dhabaan GN, Abubakar S, Cerqueira GM, Al-Haroni M, Pang SP, Hassan H. Imipenem treatment induces expression of important genes and phenotypes in a resistant Acinetobacter baumannii isolate. Antimicrob Agents Chemother. 2015;60(3):1370–1376. |
Supplementary materials
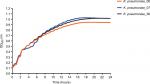  | Figure S2 The growth curves of the K. pneumoniae strains. Abbreviation: K. pneumoniae, Klebsiella pneumoniae. |
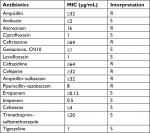  | Table S1 Antibiotics susceptibility profile of the KP_06 strain. Abbreviations: MIC, minimum inhibitory concentration; R, resistant; S, susceptible. |
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.







