Back to Journals » Infection and Drug Resistance » Volume 7
Virulence and antimicrobial resistance of Escherichia coli isolated from Tigris River and children diarrhea
Authors Ibrahim I, Al-Shwaikh R , Ismaeil M
Received 6 July 2014
Accepted for publication 25 August 2014
Published 26 November 2014 Volume 2014:7 Pages 317—322
DOI https://doi.org/10.2147/IDR.S70684
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Suresh Antony
Israa AJ Ibrahim, Rana M Al-Shwaikh, Mahmoud I Ismaeil
Department of Biology, College of Education for Pure Science, Ibn Al-Haitham, University of Baghdad, Baghdad, Iraq
Objective: To investigate the virulence factors including hemolysin production, β-lactamase production, and biofilm formation. Antimicrobial resistance and plasmid content of 20 Escherichia coli isolates obtained from feces and Tigris water were screened.
Methods: Ten clinical and ten environmental E. coli isolates were collected from children diarrhea and swim areas on Tigris River in Baghdad city, Iraq, respectively. The bacterial isolates were identified by cultural characteristics, Gram stain, biochemical tests, and screened for the presence of E. coli O157:H7 serotype. Bacterial E. coli isolates were investigated for hemolysin production, biofilm formation, and β-lactamase production. Antibiotics susceptibility and plasmid content were determined.
Results: A total of ten clinical and ten water E. coli isolates were studied. Results showed that all E. coli isolates give negative results for latex O157:H7. Virulence factors analysis showed that 6/10 water isolates and 2/10 clinical isolates were hemolytic, 5/10 water isolates and 3/10 clinical isolates were biofilm formation, and 7/10 water isolates and 4/10 clinical isolates were β-lactamase producer. Antibiotics profile showed that all bacterial isolates were multidrug resistant. All E. coli isolates (100%) were resistant to carbenicillin, cefodizime, imipenem, and piperacillin. The plasmid DNA analysis showed that all E. coli isolates contained plasmid with molecular weight range between 4.507 kbp and 5.07 kbp, but clinical isolates contained multiple small and mega plasmids.
Conclusion: Our study revealed that E. coli isolates from river water exhibit a higher level of hemolysin production, β-lactamase production, and biofilm formation than feces isolates may be due to long adaptation. On the other hand, clinical E. coli isolates from feces showed higher level of antibiotic resistance and have multiple plasmids.
Keywords: E. coli, hemolysin, β-lactamase, biofilm, multidrug resistance, plasmid
Introduction
Escherichia coli is a gram-negative bacteria belonging to the Enterobacteriaceae family, short bacilli, non-spore forming, facultative anaerobic, and it is grown on a simple media.1 E. coli is a major component of the human normal intestinal flora. Among the intestinal pathogens, there are six well-described categories: enteropathogenic E. coli (EPEC), enterohemorrhagic E. coli (EHEC), enterotoxigenic E. coli, enteraggregative E. coli, enteroinvasive E. coli, and diffusely adherent E. coli.2 There are several virulence factors that contribute to E. coli pathogenicity, such as pilli, enterotoxins (LT, ST), shiga-like toxins, endotoxin (lipopolysaccharide), hemolysin, aerobactin, cytonecrotizing factor, intimin, and biofilm formation.3,4 Plasmid profile analysis is useful in determining the epidemical strain in outbreaks caused by multiple species: Escherichia, Klebsiella, Pseudomonas, Serratia, Streptococcus, and so on.5 The adherence plasmid of EPEC strain is associated with the ability of EPEC to adhere to tissue culture cells in a pattern called localized adherence.6,7 EHEC also carry a large plasmid (91.2 kbp) that is associated with the presence of fimbriae.8 Plasmids allow the movement of genetic material, including antimicrobial resistance genes, between bacterial species and genera.9 The widespread development of resistance to several different antibiotics is generally as a result of lateral or horizontal gene transfer.10 Many studies have demonstrated that plasmid transfer between bacteria occurs in diverse environments. Various diverse phenotypic characteristics are encoded by plasmids; these include antibiotic and metal resistance, degradation of complex organic compounds, production of enterotoxins and colicins, and the production of restriction enzymes.11 Most of the transfers are described using bacteria from the same group or ecological niche.12 Plasmids size range from 1 kbp to 2,000 kbp resistance plasmids code for enzymes that can inactivate antibiotics, prevent the uptake of an antibiotic, or pump out the particular antibiotic.13 Gram-negative bacilli causing nosocomial infections are usually multiple-antibiotic resistant, resulting in significant problems for treatment.14 The aim of this study is comparing virulence factors and plasmid content among E. coli isolates from clinical and environmental sources.
Materials and methods
Specimens
In this study, 20 E. coli isolates were analyzed, ten E. coli isolates were obtained of the Tigris River water in Baghdad city and the other ten bacterial isolates were collected from children diarrhea from Children Hospital Center in Baghdad city. Samples collections were carried out from May to July 2013.
Isolation and identification of E. coli
Membrane filter technique
A known volume (30 mL) of water samples collected from swim areas on the Tigris River in Baghdad city was filtered through sterile 47 mm diameter membrane filters with 0.45 μm pores.15 The membrane filters were placed on the surfaces of eosin methylene blue agar plates for isolation of enteric bacteria and on MacConkey agar plates for isolation of lactose and nonlactose-fermenting bacteria.
E. coli isolation from clinical samples
Ten E. coli isolates were collected from children (aged under 4 years) diarrhea. Fecal samples were cultured on MacConkey agar and eosin methylene blue agar plates.16
Biochemical tests
Enteric bacteria isolated on respective selective and differential media were identified on the basis of colonial characteristics, Gram stain and biochemical tests, IMViC, Urea, and Kligler Iron Agar.16
Latex O157:H7
All E. coli isolates were investigated with latex O157:H7 (Well Colex) slide agglutination test.
Detection of hemolysin production
The E. coli isolates tested for blood hemolysis were streaked on blood agar plates containing 5% (v/v) human blood and incubated aerobically at 37°C for 24 hours. The clear zones around the growth colonies indicate a positive reaction.17
Detection of β-lactamase production
All E. coli isolates were tested for production of β-lactamase enzyme using iodometeric test method.18
Detection of biofilm formation
The ability of E. coli isolates to colonize abiotic surface was investigated by using Christensen et al’s method.19 The E. coli isolates were cultivated in tubes with Tryptone soy broth and incubated aerobically at 37°C for 48 hours and thereafter the culture tubes were emptied carefully and stained with 1% crystal violet solution for 30 minutes, and then the tubes were rinsed with distilled water and left to dry at room temperature. Results were compared with negative control, and biofilm formations as a layer at the internal wall of the tubes noted by the naked eye indicate a positive result.
Antibiotic sensitivity test
Antibiotic susceptibility profiles of E. coli isolates were determined by the standard Kirby–Bauer disk diffusion method.20 The antibiotics with their respective disk concentrations are as follows: amoxicillin–clavulanic (30 μg), carbenicillin (100 μg), cefodizime (30 μg), chloramphenicol (30 μg), gentamicin (10 μg), imipenem (10 μg), norfloxacin (10 μg), piperacillin (100 μg), rifampicin (5 μg), and pefloxacin (5 μg). Bacterial cultures suspension equivalent of 0.5 tube McFarland turbidity standards were spread on Mueller-Hinton agar plates using sterile swabs and incubated aerobically at 37°C for 24 hours, and then the diameter of inhibition zones around antibiotic disks were measured. Results were expressed susceptible or resistant according to the criteria recommended by the Clinical and Laboratory Standards Institute.21
Plasmid DNA isolated procedure
All E. coli isolates were screened for plasmid content by the alkaline method of Brinboim and Doly22 and separated on a 1% agarose, at 50 vol for 1 hour and 1.5 hours. The DNA bands were visualized and photographed under UV light after the gel had been stained with ethidium bromide.
DNA purity
DNA purity was estimated by NanoDrop method (ACT gene, USA). The absorbance at 260 nm (A260) and at 280 nm (A280) for DNA was measured to check its purity. The ratio of A260/A280 was found to be between 1.65 and 1.84.
Results and discussion
In this study, ten E. coli isolates obtained from the Tigris River in Baghdad city and ten E. coli isolates collected from children diarrhea from Children Hospital Center in Baghdad city were tested for hemolysin production, β-lactamase production, biofilm formation, antibiotic sensitivity, and plasmid content.
Latex O157:H7
Results showed that all E. coli isolates investigated with latex O157:H7 slide agglutination test gave a negative reaction. Chigor et al23 found that rate of E. coli O157 in surface waters was 2.2% and its prevalence in children with diarrhea was 5.4%. Baqir et al24 revealed that E. coli O157:H7 was isolated from four (1.14%) children with diarrhea.
Hemolysin production
The hemolytic toxin α-hemolysin belongs to the RTX toxin family25 and is an important virulence factor produced by several strains of E. coli. It is involved in human diseases such as urinary tract infections (UTIs), peritonitis, meningitis, and septicemia.26 Results illustrated in Figure 1 showed that eight (40%) E. coli isolates (six isolated from river and two isolated from children diarrhea) were producing hemolysin, while the remaining 12 (60%) isolates showed γ-hemolysis (no hemolysis) and were considered as non-hemolysin producers. Martinez-Martinez et al27 found that 56 (27.3%) of clinical E. coli isolates were hemolytic. Al-Chalabi28 recorded that 57.1% of uropathogenic E. coli produced hemolysin and Abdel Rahman29 revealed that 70.5% of E. coli isolated from chronic UTIs were hemolytic. The difference percentage of hemolysin production by E. coli isolates may be due to source of blood, type of hemolysin produced, source of bacteria, and screening method.
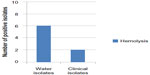  | Figure 1 Number of E. coli isolates producing hemolysin. |
β-lactamase enzyme production
Extended spectrum β-lactamases have emerged as a major threat worldwide with limited treatment options.30 The results showed that eleven (55%) E. coli isolates (seven isolated from river and four isolated from children diarrhea) were β-lactamase enzyme producers (Figure 2). This result disagreed with the result recorded by Panus et al,31 who found that 21 (14.66%) aquatic E. coli isolates (seven isolated from drinking water and 14 from sea water) were positive for β-lactamase test. Results illustrated in Figure 3 showed that all E. coli (100%) isolates were resistant to β-lactam drugs, such as carbenicillin, piperacillin, cefodizime, and imipenem. In gram-negative bacteria, a variety of plasmid-determined β-lactamases can hydrolyze ampicillin, carbenicillin, cephalothin, and related drugs.32,33 The “newer β-lactamases” that consist of plasmid-mediated AmpC β-lactamases (eg, CMY types), extended spectrum β-lactamase (eg, CTX-M types), and carbapenemases (eg, NDM) are important causes of resistance to β-lactam antibiotics among extraintestinal pathogenic E. coli.34
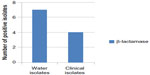  | Figure 2 Number of E. coli isolates producing β-lactamase. |
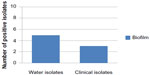  | Figure 3 Number of E. coli isolates producing biofilm. |
Biofilm formation
The biofilm formation is considered to be a two-step process in which the bacteria first adhere to a surface mediated by capsular antigen or flagellar antigen followed by multiplication to form a multilayered biofilm, which is associated with production of exopolysaccharide matrix. The ability of bacteria to form biofilms helps them to survive hostile conditions within host and is considered to be responsible for chronic or persistent infections.35
Five of the ten water E. coli isolates and two of the ten clinical E. coli isolates showed biofilm formation (Figure 3). Al-Chalabi28 found that 90% of E. coli isolated from UTI were producing biofilm. There are complexities of genetic and environmental effectors of the biofilm phenotype within the species E. coli.36
Antibiotic sensitivity
There was a high correlation between water isolates and clinical isolates of different sources in a pattern of antibiotic resistance, which implies that the water sources might have been contaminated with mixed contaminants originating from human and animal excreta.37 All E. coli isolates showed multidrug resistance (Figure 4). Overall resistance was most frequently observed with antibiotics carbenicillin, cefodizime, imipenem, and piperacillin (100%) for both clinical and water E. coli isolates; chloramphenicol showed 100% for clinical isolates and 80% for river isolates; norfloxacin showed 100% for clinical isolates and 90% for water isolates; gentamicin showed 90% for clinical isolates and 80% for water isolates; amoxicillin–calvulanic showed 90% for clinical isolates and 70% for water isolates; and pefloxacin showed 90% for clinical isolates and 50% for water isolates; water isolates showed 90% resistance to rifampicin and clinical isolates revealed 80% resistance to rifampicin. Other researchers revealed that multidrug resistance (MDR) was higher among aquatic isolates than the clinical isolates.38 The presence of antibiotic-resistant bacteria in water and surface water is a subject of public health concern; this leads to a real risk factor of acquiring such bacteria from the environments.39
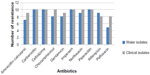  | Figure 4 Number of E. coli isolates resistant for antimicrobial agents. |
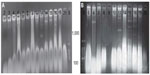
Plasmid DNA
All E. coli isolates had plasmids bands that ranged from 4.507 kbp to 5.07 kbp (Figure 5). Loss of plasmids (>2.1 kbp) due to treatment with sodium dodecyl-sulfate correlated with loss of resistance to antibiotics, suggesting that the observed MDR was plasmid mediated.38 Our results may be referring to the main β-lactamase production and MDR to plasmids range 4.507–5.07 kbp.
Clinical isolates (Number 11–20 isolates) showed multiple plasmids more than 1,000 bp (Figure 6). These multiple copies of plasmid bands might have resulted from covalently close circular, open circular, or linear forms of the same plasmid. There are several types of E. coli virulence plasmids, including those essential for the virulence of enterotoxigenic E. coli, enteroinvasive E. coli, EPEC, EHEC, enteroaggregative E. coli, and extraintestinal pathogenic E. coli.40 It has been suggested that special pathogenicity is due to small and large plasmids; some researcher revealed that 212.8 kbp plasmid was significantly associated with enteroinvasiveness41 and EHEC carry a large plasmid (91.2 kbp) that is associated with the presence of fimbriae.8
Disclosure
The authors report no conflicts of interest in this work.
References
Brooks GF, Carroll KC, Butel JS, Morse SA, Mietzner TA, Jawetz M. Adelberg’s, Medical Microbiology. 25th ed. USA: McGraw-Hill Companies, Inc.; 2010:213–225. | |
Kaper JB, Nataro JP, Mobley HLT. Pathology Escherichia coli. Nat Rev Microbiol. 2004;2:123–140. | |
Blanc M, Blanco JE, Abalia L, Alonso MP, Blanco J. Virulence factors and O groups of Escherichia coli isolates from patients with acute pyelonephritis, cystitis and asymptomatic bacteriuria. Eur J Epidemiol. 1996;12:191–198. | |
Wang G, Clark CG, Rodgers FG. Detection in Escherichia coli of the genes encoding the major virulence factors, the genes defining the O157: H7 serotype and components of the type 2 shiga toxin family by multiplex PCR. J Clin Microbiol. 2002;40(10):3613–3619. | |
Wachsmuth K. Molecular epidemiology of bacterial infections: examples of methodology and of investigations of outbreaks. Rev Infect Dis. 1986;8:682–692. | |
Baldini MM, Kaper JB, Levine MM, Candy DCA, Moon HW. Plasmid-mediated adhesion in enteropathogenic Escherichia coli. J Pediatr Gastroenterol Nutr. 1983;2:534–538. | |
Nataro JP, Scaletsky ICA, Kaper JB, Levine MM, Trabulsl LR. Plasmid-mediated factors conferring diffuse and localized adherence of enteropathogenic Escherichia coli. Infect Immun. 1985;48:378–383. | |
Karch H, Heesemann J, Laufs R, O’Brien AD, Tacket CO, Levine MM. A plasmid of enterohemorrhagic Escherichia coli O157:H7 is required for expression of a new fimbrial antigen and for adhesion to epithelial cells. Infect Immun. 1987;55:455–461. | |
Sherley M, Gordon DM, Collignon PJ. Evolution of multi-resistance plasmids in Australian clinical isolates of Escherichia coli. Microbiology. 2004;150:1539–1546. | |
Woo PC, To APC, Lau SK, Yuen KY. Facilitation of horizontal transfer of antimicrobial resistance by transformation of antibiotic-induced cell-wall-deficient bacteria. Med Hypotheses. 2003;61(4):503–508. | |
Maniatis T, Fritsch EF, Sambrook J. Molecular Cloning. A Laboratory Manual. Cold Spring, USA: Cold Spring Harbor Laboratory; 1982:1–545. | |
Kruse H, Serum H. Transfer of multiple drug resistance plasmids between bacteria of diverse origins in natural environments. App Environ Microbiol. 1994;60(11):4015–4021. | |
Madigan M, Martinko J, Parker J. Brock Biology of Microorganisms. 10th ed. Upper Saddle River, NJ, USA: Prentice Hall; 2003. | |
Newman MC, Scheuren-Portocarrero SM. Multiple Antibiotic Resistances: What is the Cure? Kentucky: University of Kentucky, Courtesy of Altech Inc.; 2007. | |
Hleyn J, Bicknell M. Microbiology Experiments: A Health Science Perspective. 5th ed. McGraw Hill Companies, Inc.; 2007. | |
Forbes BA, Sahm DF, Weissfeld AS. Bailey and Scotts, Diagnostic Microbiology. 11th ed. Missouri: Mosby; 2002. | |
Senior BW, Hughes C. Production and properties of hemolysin from clinical isolates of Proteus. J Med Microbiol. 1987;24:17–25. | |
Livermore DM, Brown DFJ. Detection of β-lactamase-mediated resistance. J Antimicrob Chemother. 2001;48:59–64. | |
Christensen GD, Bisno AL, Parisi JT, McLaughlin B, Hester MG, Luther RW. Nosocomial septicemia due to multiply antibiotic resistant Staphylococcus epidermidis. Ann Intern Med. 1982;96:1–10. | |
Bauer AW, Kirby WMM, Sherris JC, Truck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1966;43:493–496. | |
Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Second Informational Supplement. CLSI Document M 100-S22. Wayne, PA: Clinical and Laboratory Standards Institute; 2012. | |
Brinboim HC, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acid Res. 1979;7(6):1513–1523. | |
Chigor VN, Umoh VJ, Smith SI, Igbinosa EO, Okoh AI. Multidrug resistance and plasmid patterns of Escherichia coli O157 and other E. coli isolated from diarrhoeal stools and surface waters from some selected sources in Zaria, Nigeria. Int J Environ Res Public Health. 2010;7:3831–3841. | |
Baqir HE, Al-Hashemi AB, Althwani NA. Microbiological study of Escherichia coli O157:H7 isolated from bloody diarrhea in children under ten years old. Iraqi J Sci. 2008;49(1):90–94. | |
Coote JG. Structural and functional relationships among the RTX toxin determinants of gram-negative bacteria. FEMS Microbiol Rev. 1992;88:137–162. | |
Cavalieri S, Bohach GA, Synder I. Escherichia coli hemolysin: characteristics and probable role in pathogenicity. Microbiol Rev. 1984;48:326–334. | |
Martinez-Martinez L, Ferandez F, Perea EJ. Relationship between haemolysis production and resistance to fluoroquinolones among clinical isolates of Escherichia coli. J Antimicrob Chemother. 1999;43:277–279. | |
Al-Chalabi RN. Relation between hemolysin production and biofilm formation by uropathogenic Escherichia coli, MSc Thesis, AL-Nahrain University, Iraq, 2004. | |
Abdel Rahman MII. Study of some bacteriological and immunological parameters in chronic urinary tract infection, PhD Thesis, Al-Mustansiriya University, Iraq, 2006. | |
Goyal A, Prasad KN, Prasad A, Gupta S, Ghoshal U, Ayyagari A. Extended spectrum β-lactamases in Escherichia coli and K. pneumoniae and associated risk factors. Indian J Med Res. 2009;129:695–700. | |
Panus E, Chifiriuc M-CB, Bucur M, et al. Virulence, pathogenicity, antibiotic resistance and plasmid profile of Escherichia coli strains isolated from drinking and recreational waters. In: 17th European Congress of Clinical Microbiology and Infectious Diseases and 25th International Congress of Chemotherapy; March 31, 2008–April 3, 2007. | |
Medeiros AA. β-Lactamases. Br Med Bull. 1984;40:18–27. | |
Sykes RB, Matthew M. The β-lactamases of gram-negative bacteria. J Antimicrob Chemother. 1976;2:115–157. | |
Pitout JDD. Extraintestinal pathogenic Escherichia coli: a combination of virulence with antibiotic resistance. Front Microbiol. 2012;3(9):1–7. | |
Costerton IW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. | |
Karen AK, Bjarke MK, Ellen LZ. In vitro biofilm formation of commensal and pathogenic Escherichia coli strains impact of environmental and genetic factors. J Bacteriol. 2006;188(10):3572–3581. | |
Abera A, Bahiru ESA, Ayele AK. The prevalence of antibiotic resistant Escherichia coli isolates from fecal and water sources. Acad J Microbiol Res. 2013;1(1):001–010. | |
Chigor VN, Umoh VJ, Smith SI, Igbinosa EO, Okoh AI. Multidrug resistance and plasmid patterns of Escherichia coli O157 and other E. coli isolated from diarrhoeal stools and surface waters from some selected sources in Zaria, Nigeria. Int J Environ Res Public Health. 2010;7:3831–3841. | |
Akturk S, Dincer S, Torogiu S. Determination of microbial quality and plasmid-mediated multidrug resistant bacteria in fountain drinking water sources in Turkey. J Environ Biol. 2012;33:1127–1136. | |
Johnson TJ, Nolan LK. Pathogenomics of the virulence plasmids of Escherichia coli. Microbiol Mol Biol Rev. 2009;73(4):750–774. | |
Harris JR, Wachsmuth IK, Davis BR, Cohen ML. High-molecular-weight plasmid correlates with Escherichia coli enteroinvasiveness. Infect Immun. 1982;37(3):1295–1298. |
 © 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.