Back to Journals » OncoTargets and Therapy » Volume 13
Variants of MIR137HG Genes are Associated with Liver Cancer Risk in Chinese Li Population
Authors Wang C, Zhuang X, Xu J, Dai Z, Wu W, Zhang C, Lin S, Chen S, Lin H, Tang W
Received 1 August 2019
Accepted for publication 11 February 2020
Published 28 February 2020 Volume 2020:13 Pages 1809—1818
DOI https://doi.org/10.2147/OTT.S225669
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Jianmin Xu
Chaoying Wang, 1,* Xiaohong Zhuang, 1,* Junnv Xu, 1, 2,* Zhisheng Dai, 3 Weixiong Wu, 4 Chengsheng Zhang, 1 Shu Lin, 1 Sehong Chen, 3 Haifeng Lin, 1 Wenjun Tang 1
1Department of Medical Oncology, The Second Affiliated Hospital of Hainan Medical University, Haikou, Hainan, 570311, People’s Republic of China; 2Hainan Medical University, Haikou, Hainan 571199, People’s Republic of China; 3Department of Medical Oncology, The Second People’s Hospital of Hainan Province, Wuzhishan, Hainan 572200, People’s Republic of China; 4Intensive Care Medicine 1 District, The Second Affiliated Hospital of Hainan Medical College, Haikou, Hainan 570311, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Haifeng Lin; Wenjun Tang #48, Baishuitang Road, Haikou 570311, Hainan, People’s Republic of China
Tel/Fax +8613322060949;
+8613322060949
Email [email protected]; [email protected]
Background: Liver cancer (LC) is the sixth most common cancer and the second leading cause of cancer mortality worldwide, and its incidence rate is high in China.
Methods: In this study, we aimed to investigate the contribution of MIR137HG (MIR137 Host Gene) polymorphisms to LC risk in a case–control study with 432 LC patients and 430 healthy controls. A logistic recession model was used to evaluate the effects of candidate single nucleotide polymorphisms (SNPs) on LC risk. HaploReg v 4.1 database was conducted to predict the potential functionality of SNPs.
Results: The results revealed that rs17371457 and rs7554283 in the MIR137HG gene were correlated with an enhanced LC risk under the allele (P = 0.001 and P = 0.043, respectively) and genetic models (P < 0.05). When the sample was stratified by gender and age, statistically significant associations were found. Rs9440302, rs17371457 and rs7554283 were associated with an increased the risk of LC among individuals aged > 55 years (P < 0.05); rs17371457 was related to higher LC risk in males (P < 0.05). Similarly, the haplotype AG constituted by rs12333983 and rs3735451 significantly increased LC risk in Chinese Li population (P = 0.043). Six SNPs distributed in MIR137HG were successfully predicted as regulatory SNPs with different biological functions.
Conclusion: Our research firstly showed that MIR137HG gene polymorphisms were implicated in LC susceptibility among Chinese Li population.
Keywords: liver cancer, genetics polymorphisms, MIR137HG, susceptibility
Background
Liver cancer (LC) is the sixth most common cancer and the second largest cause of cancer mortality worldwide, with nearly 800,000 cases in 2012.1 The highest incidence rates of LC were observed in Eastern Asia, South-Eastern Asia, Northern Africa and Southern Africa, with China accounting for about 50% of all cases.2 LC is usually caused by underlying diseases, including hepatitis B virus (HBV), mycotoxins, poor diet and inactivity.1 However, there is a startling lack of treatments on LC.
MiRNA is regarded as the critical elements in cancer development and involved in major cellular processes of cancer, such as cell differentiation, proliferation, migration, and apoptosis.3–5 Further studies have shown that miRNAs can be used as tumor initiation factors, invasion and metastasis factors, tumor suppressor factors, etc. Hence, MiRNA may have a potential role in the development of new cancer control methods.4,6
MIR137HG (MIR137 Host Gene) is an RNA Gene and is affiliated with the miRNA class. Currently, researches have shown that the promoter of MIR137HG is highly methylated in colorectal cancer,7 malignant pleural mesothelioma,8 and oral squamous cell carcinoma.9 MIR137 is known to be a tumor suppressor, and the increased methylation level of MIR137HG in malignant pleural mesothelioma cell lines and colorectal cancer affects the occurrence and development of corresponding cancers.7 However, there are no previous studies have investigated the association of LC risk and MIR137HG polymorphisms.
Hence, a case-control study was performed in 432 LC patients and 430 healthy controls to evaluate the possible association of LC and MIR137HG polymorphisms among Chinese Li population. The investigation was expected to further deepen our understanding regarding the pathogenesis of LC and serve as non-invasive biomarkers for LC.
Materials and Methods
Ethics Statement
Our research was approved by the Ethics Committee of the Second Affiliated Hospital of Hainan Medical College (Ethics number: HZ2015-11), and all procedures were conducted in accordance with the ethical standards of the Declaration of Helsinki and following national and international guidelines. Written informed consent was obtained from each participant after the study was fully explained.
Study subjects862 individuals, consisting of 430 healthy people and 432 patients with LC, were recruited from the Second Affiliated Hospital of Hainan Medical College, Hainan Province, China. All LC patients were newly diagnosed and had not received relevant antitumor therapy, such as surgical treatment, radiotherapy, chemotherapy and biotherapy. They were diagnosed with liver cancer by routine examination, imaging (such as B-ultrasonic wave, CT and nuclear magnetic resonance) and histopathological examination. Our case group also excluded patients with the following diseases: liver diseases like hepatitis and liver cirrhosis induced by diabetes, fatty liver, metabolic disorder and vascular disease, primary and secondary biliary cirrhosis, recurrent liver cancer, metastatic liver cancer, hepatitis A, C, D, and E, autoimmune hepatitis, etc. Control subjects, without any history of tumor or chronic diseases, were healthy individuals selected from the physical examination center of the Second Affiliated Hospital of Hainan Medical College, Hainan Province, China. All the participants were Li nationality from Hainan Province of China and they have no relationship with each other.
SNP Selection and Genotyping
According to the 1000 Genomes Project (http://www.1000genomes.org/) and dbSNP (https://www.ncbi.nlm.nih.gov/projects/SNP/) database, five label single nucleotide polymorphisms (SNPs) (rs12138817, rs9440302, rs1198574, rs17371457 and rs7554283) with minor allele frequency (MAF) > 5% were finally selected to evaluate the effect of MIR137HG polymorphisms on LC susceptibility.
Peripheral blood samples from all participants were collected in tubes coated with EDTA and were stored at 80°C. Following the manufacturer’s guidelines, genomic DNA was extracted from participants’ peripheral blood samples using the GoldMag whole blood genomic DNA purification kit (GoldMag Co. Ltd., Xi an, China). DNA concentration was measured with NanoDrop 2000 spectrophotometer (Thermo Scientific, Waltham, MA, USA).10 The primers for the amplification reactions were designed using the Agena Bioscience Assay Design Suite V2.0 software to perform (Agena Bioscience, San Diego, CA, USA, https://agenacx.com/online-tools/).11 Primers used for this study were listed in Supplementary Table S1. The MassARRAY iPLEX platform and Agena Bioscience TYPER version 4.0 software were used for SNP genotyping and data analysis, respectively.
SNP Functional Evaluation
HaploReg v4.1 (https://pubs.broadinstitute.org/mammals/haploreg/haploreg.php) was used to predict the potential functions of SNPs in MIR137HG gene.
Statistical Analysis
SPSS software package (version 20.0; SPSS Inc., Chicago, IL, USA) was adopted to process data, and P < 0.05 was regarded as statistically significant.12,13 Hardy–Weinberg equilibrium (HWE) P values obtained from exact tests14 were used to evaluate whether the control group meets HWE. The distributions of genotype and allele frequencies between cases and controls were compared by χ2 tests. The genotype-specific risks were estimated as odds ratios (ORs) and 95% confidence interval (CI) based on logistic regression model analysis.15 Stratified analyses were also performed to assess the relationship between each SNP and the risk of LC in different subgroups. Finally, linkage disequilibrium (LD) analyses and haplotype analyses were performed using the SHEsis software (http://analysis.bio-x.cn/myAnalysis.php).
Results
Participant Characteristics
The basic demographic characteristics of study population are described in Table 1. A total of 432 LC patients (344 males and 88 females, age at diagnosis: 55.09 ± 11.59 years) and 430 controls (342 males and 88 females, age at diagnosis: 55.22 ± 10.73) were enrolled in our present study. There was no statistical difference in gender and age distribution between cases and control groups (P > 0.05).
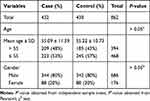 |
Table 1 Distributions of Age and Gender in LC Cases and Healthy Controls |
Basic Information and Preliminary Statistics of the Selected SNPs
Basic information containing SNP ID, chromosomal position, alleles, MAF distribution, P-HWE, call rate, ORs and 95% CIs of all the candidate SNPs were demonstrated in Table 2. The call rate for all SNPs was >99.00% among the LC cases and the controls. All candidate SNPs were in accordance with HWE (P-HWE > 0.05), indicating good sample selection. From the five SNPs, the minor allele T of rs17371457 and the minor allele G of rs7554283 in the MIR137HG were significantly correlated with an enhanced the risk of LC (OR = 1.60, 95% CI = 1.01–1.47, P = 0.001; OR = 1.22, 95% CI = 1.01–1.47, P = 0.043, respectively).
 |
Table 2 Basic Information and Allele Frequency of the Selected SNPs in MIR137HG Gene |
Associations Between Genotype Frequencies and LC Risk
Furthermore, the correlations between polymorphisms and LC susceptibility were analyzed based on multiple inheritance modes (codominant, dominant, recessive, and log-additive models) using logistic tests. As shown in Table 3, rs17371457 was found to be correlated with an increased the risk of LC in the codominant model (C/T vs C/C: OR = 1.41, 95% CI = 1.01–1.96, P = 0.044; T/T vs C/C: OR = 5.14, 95% CI = 1.46–18.04, P = 0.011), dominant model (OR = 1.54, 95% CI = 1.12–2.13, P = 0.008), recessive model (OR = 4.78, 95% CI = 1.36–16.76, P = 0.015), and log-additive model (OR = 1.58, 95% CI = 1.18–2.11, P = 0.002). Rs7554283 was significantly correlated with an enhanced LC risk in the recessive model (OR = 1.39, 95% CI = 1.02–1.90, P = 0.038) and log-additive model (OR = 1.22, 95% CI = 1.01–1.47, P = 0.042). No significant correlation was observed between the remaining SNPs (rs12333983, rs3735451, rs4646437 and rs2246709) and LC risk.
 |
Table 3 Significant Genetic Variants in MIR137HG Gene Associated with the Susceptibility of LC in Chinese Li Population |
Stratification Analysis by Age and Gender
According to the average age, stratified analyses regarding the impact of SNPs on the risk of LC are summarized in Table 4. Among those aged over 55 years, the genotype A/G-G/G of rs17371457 was associated with an increased LC risk in the codominant model (OR = 1.56, 95% CI = 1.01–2.42, P = 0.047). Rs17371457 significantly increased LC risk in the codominant model (CT vs CC: OR = 1.76, 95% CI = 1.05–2.93, P = 0.031; TT vs CC: OR = 8.32, 95% CI = 1.03–67.41, P = 0.047), dominant model (OR = 1.98, 95% CI = 1.20–3.25, P = 0.007), and log-additive model (OR = 1.97, 95% CI = 1.26–3.08, P = 0.003). We also found that rs7554283 was correlated with an increased risk of LC in the codominant model (G/G vs C/C: OR = 1.82, 95% CI = 1.02–3.23, P = 0.042) and log-additive model (OR = 1.35, 95% CI = 1.01–1.80, P = 0.042). No significant association was found between the five SNPs loci in MIR137HG and LC risk at age ≤50 years.
 |
Table 4 Relationship of MIR137HG Polymorphisms with LC Risk Stratified by Age |
Stratified analyses by gender were also conducted. As shown in Table 5, the results revealed that rs17371457 was correlated with an enhanced risk of LC in males under the codominant model (C/T vs C/C: OR = 1.52, 95% CI = 1.01–1.80, P = 0.042; T/T vs C/C: OR = 14.61, 95% CI = 1.90–112.50, P = 0.010), dominant model (OR = 1.73, 95% CI = 1.20–2.50), P = 0.003), recessive model (OR = 13.41, 95% CI = 1.75–103.10, P = 0.013), log-additive model (OR = 1.81, 95% CI = 1.30–2.52, P < 0.001) and allele model (OR = 1.85, 95% CI = 1.33–2.58, P < 0.001). In the subgroup of females, we did not find a significant association between candidate SNPs in MIR137HG and LC risk.
 |
Table 5 Relationship of MIR137HG Polymorphisms with LC Risk Stratified by Gender |
Associations Between Haplotype Analyses and LC Risk
Finally, we studied the LD and haplotype analysis of candidate SNPs. As shown in Figure 1, the LD block in MIR137HG gene was constituted of two SNPs (rs9440302 and rs1198574). The frequency distribution of haplotypes in two groups and its association with LC risk were presented in Table 6. Haplotype AG in the MIR137HG gene was found to prominently increase the risk of LC (OR = 1.22, 95% CI = 1.01–1.47; P = 0.043), other haplotypes did not display the correlativity.
 |
Table 6 Haplotype Frequencies of MIR137HG Gene and the Association with LC Risk |
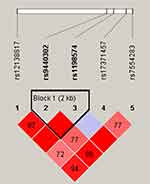 |
Figure 1 Haplotype block map for six SNPs in MIR137HG Gene. |
SNP Functional Evaluation
In order to explore the regulatory roles of the statistically significant variants, we predicted the function of them using HaploReg v4.1 database. Six SNPs in MIR137HG were successfully predicted as regulatory SNPs with different biological functions (Supplementary Table S2).
Discussion
The miRNA variants have been recognized as major contributors to the pathogenesis and development of cancers. In this study, we aimed to elucidate the relationship between MIR137HG gene polymorphisms and LC susceptibility in Chinese Li population. The results suggested that rs17371457 and rs7554283 in the MIR137HG gene were correlated with an enhanced LC risk. After stratifying the sample by gender and age, correlation were found between higher risk of LC and MIR137HG polymorphisms (rs9440302, rs17371457 and rs7554283) at age >55 years. Rs17371457 was related to higher LC risk in males. Similarly, the haplotype AG significantly increased LC risk in Chinese Li population. As far as we know, this is the first study to show the association of MIR137HG polymorphisms and LC susceptibility in Chinese Li population.
MiRNAs are involved in the major cellular processes of cancer, and MIR137HG belongs to the miRNA class. Previous studies showed that MIR137HG gene polymorphisms affect the gray matter structure in patients with schizophrenia, and it was also related to the volume of corpus callosum.16,17 At the same time, it has been reported that the methylation level of MIR137HG promoter was increased in a variety of cancers and was related to the expression level of MIR137. Morandi et al believed that methylation level was the key factor for early detection of oral squamous cell carcinoma (OSCC), and found that MIR137HG was hypermethylated in OSCC and high-grade squamous intraepithelial lesion (HGSIL).9 Johnson et al observed that MIR137HG was methylated in malignant pleural mesothelioma (MPM) cell lines. Besides, MIR137 showed tumor inhibition function in MPM by targeting YBX1, which was dysregulated in MPM because of Copy number variations (CNV) and hypermethylation of the MIR137HG promoter.8 Li et al showed that HSF1 recruited DNMT3a to inhibit the promoter methylation of lncRNA MIR137HG by targeting GLS1 mRNA, and further reduced the expression of MIR137HG-MIR137, and HSF1 stimulated GLS1-dependent mTOR activation to promote colorectal carcinogenesis.7
Furthermore, MIR137HG is the host gene of MIR137. It is reported that MIR137 was a tumor suppressor by inhibiting cancer proliferation, cell-cycle progression and migration,18,19 and it was lowly expressed in various cancers, such as hepatocellular carcinoma, lung cancer, colorectal cancer, gastric cancer, and cervical cancer.20–22 Additionally, studies have shown that MIR137 may serve as a prognostic marker in hepatocellular carcinoma (HCC) patients,23 and miR-137/miR-133a-TINCR pathway can be used as a promising target for tumor recurrence and prognosis in HCC patients.24 Researches have shown that MIR137 inhibits tumor growth and metastasis in HCC by targeting AKT serine/threonine kinase225 and the suppressive effects of MIR137 on HCC cell proliferation and metastasis were regulated by cell division cycle.26 MIR137 was down-regulated in HCC and inhibits HCC cell migration and invasion by targeting EZH2-STAT3 signaling pathway.27 Therefore, MIR137HG may also affect the expression of MIR137 through promoter hypermethylation, thereby affecting the metastasis and growth of LC.
Significantly, the major allele of MIR137HG risk SNPs, including rs1625579, lead to lower levels of mature MIR137 than those with the minor allele.28 A post-mortem tissue analysis also showed that the biogenesis of this miRNA may be disrupted in carriers of the rs1625579 risk genotypes.29 In our case–control study, we investigated the correlation between MIR137HG SNPs and LC risk, and the result showed that rs17371457 and rs7554283 in MIR137HG gene exhibited an increased risk of LC. Rs9440302, rs17371457 and rs7554283 also correlation with an increased LC risk at age >55 years. The rs17371457 was related to higher LC risk in males. Based on these results, we hypothesized that the allele of MIR137HG risk SNPs (s9440302, rs17371457 and rs7554283) reduced the level of mature MIR137, thereby increasing the risk of LC. Additionally, the case group in our study included patients with confirmed liver cancer, excluding patients with liver cirrhosis, chronic hepatitis B and C, NAFLD, Wilson’s disease and other liver diseases. Thus, the distribution of MIR137HG SNPs in other liver diseases (such as chronic B and C hepatitis, cirrhosis, NAFLD and Wilson’s disease), and the relationship with liver-related diseases are also unknown. The specific mechanism of MIR137HG SNPs on LC risk and the relationship between MIR137HG SNPs and other liver diseases still need further study. Several limitations of this study should not be ignored. Firstly, current study may be limited by the source of samples, which may limit statistical capacity. Secondly, the size of sample was not large enough to rule out false negative results. So, larger sample size and more ethnic groups are needed for further verification. Despite the above limitations, the results of this study provide a scientific basis for future studies on the relationship between LC and MIR137HG gene SNPs.
In general, this study firstly revealed the potential role of MIR137HG on LC susceptibility in the Chinese Li population, which provides new insights into the pathogenesis of LC. Further studies with larger samples and well-designed methods in diverse populations are necessary to confirm our results.
Acknowledgments
This work is supported by the Project of Hainan Health and Family Planning Science and Education (Grant No. 15A200102). We are grateful to the individuals for their participation in this study. We also thank the clinicians and hospital staff who contributed to the sample and data collection for this study. We would also like to thank all participants for this manuscript.
Disclosure
The authors report that they have no conflicts of interest in this work.
References
1. Gravitz L. Liver cancer. Nature. 2014;516(7529):S1. doi:10.1038/516S1a
2. Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–E386. doi:10.1002/ijc.29210
3. Tam C, Wong JH, Tsui SKW, Zuo T, Chan TF, Ng TB. LncRNAs with miRNAs in regulation of gastric, liver, and colorectal cancers: updates in recent years. Appl Microbiol Biotechnol. 2019;103(12):4649–4677.
4. Price C, Chen J. MicroRNAs in cancer biology and therapy: current status and perspectives. Genes Dis. 2014;1(1):53–63. doi:10.1016/j.gendis.2014.06.004
5. Tutar L, Ozgur A, Tutar Y. Involvement of miRNAs and Pseudogenes in cancer. Methods Mol Biol. 2018;1699:45–66.
6. Liu M, Jiang L, Guan XY. The genetic and epigenetic alterations in human hepatocellular carcinoma: a recent update. Protein Cell. 2014;5(9):673–691. doi:10.1007/s13238-014-0065-9
7. Li J, Song P, Jiang T, et al. Heat shock factor 1 epigenetically stimulates glutaminase-1-dependent mTOR activation to promote colorectal carcinogenesis. Mol Ther. 2018;26(7):1828–1839. doi:10.1016/j.ymthe.2018.04.014
8. Johnson TG, Schelch K, Cheng YY, et al. Dysregulated expression of the MicroRNA miR-137 and its target YBX1 contribute to the invasive characteristics of malignant pleural mesothelioma. J Thorac Oncol. 2018;13(2):258–272. doi:10.1016/j.jtho.2017.10.016
9. Morandi L, Gissi D, Tarsitano A, et al. CpG location and methylation level are crucial factors for the early detection of oral squamous cell carcinoma in brushing samples using bisulfite sequencing of a 13-gene panel. Clin Epigenetics. 2017;9:85. doi:10.1186/s13148-017-0386-7
10. Geng TT, Xun XJ, Li S, et al. Association of colorectal cancer susceptibility variants with esophageal cancer in a Chinese population. World J Gastroenterol. 2015;21(22):6898–6904.
11. Wang T, Chen T, Thakur A, et al. Association of PSMA4 polymorphisms with lung cancer susceptibility and response to cisplatin-based chemotherapy in a Chinese Han population. Clin Transl Oncol. 2015;17(7):564–569. doi:10.1007/s12094-015-1279-x
12. Huang CY, Xun XJ, Wang AJ, et al. CHRNA5 polymorphisms and risk of lung cancer in Chinese Han smokers. Am J Cancer Res. 2015;5(10):3241–3248.
13. Zhang T, Li X, Du Q, et al. DUSP10 gene polymorphism and risk of colorectal cancer in the Han Chinese population. Eur J Cancer Prev. 2014;23(3):173–176. doi:10.1097/CEJ.0b013e3283647408
14. Lorenzl S, De Pasquale G, Segal AZ, Beal MF. Dysregulation of the levels of matrix metalloproteinases and tissue inhibitors of matrix metalloproteinases in the early phase of cerebral ischemia. Stroke. 2003;34(6):
15. Newby AC. Matrix metalloproteinase inhibition therapy for vascular diseases. Vascul Pharmacol. 2012;56(5–6):232–244. doi:10.1016/j.vph.2012.01.007
16. He E, Lozano MAG, Stringer S, et al. MIR137 schizophrenia-associated locus controls synaptic function by regulating synaptogenesis, synapse maturation and synaptic transmission. Hum Mol Genet. 2018;27(11):1879–1891. doi:10.1093/hmg/ddy089
17. Wright C, Gupta CN, Chen J, et al. Polymorphisms in MIR137HG and microRNA-137-regulated genes influence gray matter structure in schizophrenia. Transl Psychiatry. 2016;6:e724. doi:10.1038/tp.2015.211
18. Shen H, Wang L, Ge X, et al. MicroRNA-137 inhibits tumor growth and sensitizes chemosensitivity to paclitaxel and cisplatin in lung cancer. Oncotarget. 2016;7(15):20728–20742. doi:10.18632/oncotarget.8011
19. Liang ML, Hsieh TH, Ng KH, et al. Downregulation of miR-137 and miR-6500-3p promotes cell proliferation in pediatric high-grade gliomas. Oncotarget. 2016;7(15):19723–19737. doi:10.18632/oncotarget.7736
20. Zhang H, Yan T, Liu Z, et al. MicroRNA-137 is negatively associated with clinical outcome and regulates tumor development through EZH2 in cervical cancer. J Cell Biochem. 2018;119(1):938–947. doi:10.1002/jcb.26259
21. Yang YR, Li YX, Gao XY, Zhao SS, Zang SZ, Zhang ZQ. MicroRNA-137 inhibits cell migration and invasion by targeting bone morphogenetic protein-7 (BMP7) in non-small cell lung cancer cells. Int J Clin Exp Pathol. 2015;8(9):10847–10853.
22. Smith AR, Marquez RT, Tsao WC, et al. Tumor suppressive microRNA-137 negatively regulates Musashi-1 and colorectal cancer progression. Oncotarget. 2015;6(14):12558–12573. doi:10.18632/oncotarget.3726
23. Sakabe T, Azumi J, Umekita Y, et al. Prognostic relevance of miR-137 in patients with hepatocellular carcinoma. Liver Int. 2017;37(2):271–279. doi:10.1111/liv.2017.37.issue-2
24. Li J, Gao J, Tian W, Li Y, Zhang J. Long non-coding RNA MALAT1 drives gastric cancer progression by regulating HMGB2 modulating the miR-1297. Cancer Cell Int. 2017;17:44. doi:10.1186/s12935-017-0408-8
25. Liu LL, Lu SX, Li M, et al. FoxD3-regulated microRNA-137 suppresses tumour growth and metastasis in human hepatocellular carcinoma by targeting AKT2. Oncotarget. 2014;5(13):5113–5124. doi:10.18632/oncotarget.2089
26. Gao M, Liu L, Li S, Zhang X, Chang Z, Zhang M. Inhibition of cell proliferation and metastasis of human hepatocellular carcinoma by miR-137 is regulated by CDC42. Oncol Rep. 2015;34(5):2523–2532. doi:10.3892/or.2015.4261
27. Huang B, Huang M, Li Q. MiR-137 suppresses migration and invasion by targeting EZH2-STAT3 signaling in human hepatocellular carcinoma. Pathol Res Pract. 2018;214(12):1980–1986. doi:10.1016/j.prp.2018.08.005
28. Siegert S, Seo J, Kwon EJ, et al. The schizophrenia risk gene product miR-137 alters presynaptic plasticity. Nat Neurosci. 2015;18(7):1008–1016. doi:10.1038/nn.4023
29. Guella I, Sequeira A, Rollins B, et al. Analysis of miR-137 expression and rs1625579 in dorsolateral prefrontal cortex. J Psychiatr Res. 2013;47(9):1215–1221. doi:10.1016/j.jpsychires.2013.05.021
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
