Back to Journals » OncoTargets and Therapy » Volume 12
Variability within the human TERT gene, telomere length and predisposition to chronic lymphocytic leukemia
Authors Wysoczanska B, Dratwa M, Gebura K , Mizgala J, Mazur G , Wrobel T , Bogunia-Kubik K
Received 14 December 2018
Accepted for publication 8 April 2019
Published 31 May 2019 Volume 2019:12 Pages 4309—4320
DOI https://doi.org/10.2147/OTT.S198313
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Sanjay Singh
Barbara Wysoczanska,1 Marta Dratwa,1 Katarzyna Gebura,1 Jakub Mizgala,1 Grzegorz Mazur,2 Tomasz Wrobel,3 Katarzyna Bogunia-Kubik1
1Laboratory of Clinical Immunogenetics and Pharmacogenetics, Hirszfeld Institute of Immunology and Experimental Therapy, Polish Academy of Sciences, Wroclaw 53-114, Poland; 2Department of Internal Medicine, Occupational Diseases, Hypertension and Clinical Oncology, Wroclaw Medical University, Wroclaw, 50-001, Poland; 3Department of Haematology, Blood Neoplasms and Bone Marrow Transplantation, Wroclaw Medical University, Wroclaw 50-367, Poland
Background: The human telomerase reverse transcriptase (TERT) gene encodes the catalytic subunit of telomerase that is essential for maintenance of telomere length. We aimed to find out whether variability within the TERT gene could be associated with telomere length and development of the disease in non-treated patients with chronic lymphocytic leukemia (CLL).
Materials and methods: Telomere length, rs2736100, rs2853690, rs33954691, rs35033501 single-nucleotide polymorphisms, and variable number of tandem repeats (VNTR-MNS16A) were assessed in patients at diagnosis. In addition, blood donors served as controls for the polymorphism studies.
Results: The minor rs35033501 A variant was more frequent among CLL patients than in healthy controls (OR=3.488, p=0.039). CLL patients over 60 years of age were characterized with lower disease stage at diagnosis (p=0.001 and p=0.008, for the Rai and Binet criteria, respectively). The MNS16A VNTR-243 short allele was more frequent in patients with a low disease stage (p=0.020 and p=0.028, for the Rai and Binet staging system) and also among older patients having longer telomeres (p=0.046). Patients with Rai 0–I stage were characterized with longer telomeres than those with more advanced disease (p=0.030). This relationship was especially pronounced in patients carrying the rs2736100 C allele, independently of the criteria used, ie, Binet (p=0.048) or Rai (p=0.001).
Conclusion: Our results showed that the genetic variation within the TERT gene seems to play a regulatory role in CLL and telomere length.
Keywords: telomere length, human telomerase reverse transcriptase, variable number of tandem repeats, single-nucleotide polymorphism, chronic lymphocytic leukemia
Introduction
Chronic lymphocytic leukemia (CLL) is the most common leukemia among adults. It primarily affects the elderly and occurs twice as often in males than in females.1 The disease is influenced by complex heterogeneous genetic and microenvironmental factors that can result in different clinical courses.2,3 The pathogenesis of CLL varies depending on molecular heterogeneity background, mutational load and specific genomic aberrations.4,5 Genome-wide association studies identified many susceptibility loci involved in B-cell biology and CLL development.6–8 One study revealed that the telomere/telomerase system may be impaired in the early stages of CLL.9 Also, several studies focused on the potential prognostic significance of telomere length and human telomerase reverse transcriptase (hTERT) activity in CLL.10–13
Human telomerase activity is regulated by expression of the telomerase reverse transcriptase (TERT) gene that encodes the catalytic subunit of telomerase.14 The TERT gene is located on chromosome 5p15.33 and consists of 16 exons and 15 introns spanning over 40 kb, and is essential for maintenance of telomere length by protecting chromosome ends. It has been documented that single-nucleotide polymorphisms (SNPs) within the TERT gene may affect the length of telomeres and telomerase activity, thus contributing to CLL disease susceptibility.7,8,15–17 Main processes such as transcription, alternative mRNA splicing, phosphorylation and many other changes, including mutations and gene variants of TERT, have been shown to play pivotal roles in the regulation of telomerase activity and cancer risk.18,19
Alternative splicing of hTERT mRNA is considered to be one of the most precise regulators of telomerase activity in human cells. Induction of an hTERT splicing variant was associated with increased expression of the apoptotic endonuclease EndoG, a splicing regulator. It was demonstrated that EndoG (an apoptotic endonuclease capable of destroying both DNA and RNA) induced alternative splicing of the telomerase catalytic subunit hTERT and inhibited telomerase activity in normal human CD4+ T lymphocytes.20,21
The promoter region and sequences upstream interact with both positive and negative regulators of the TERT gene through many transcriptional binding sites.22,23 It has been demonstrated that TERT promoter activity depends on variable numbers of tandem repeats (VNTRs), such as MNS16A, which constitutes a binding site for a transcription factor GATA binding protein 1 (GATA-1).24 Research on the functional significance of this genetic polymorphism showed that shorter MNS16A is related to higher TERT promoter activity.24,25 This functional polymorphism may play an important role in human longevity, disease progression and response to therapy of patients with non-Hodgkin’s B-cell lymphomas and the development of other cancers.26–29
Mechanisms underlying telomere maintenance and telomerase reactivation in leukemogenesis are currently being investigated. Increasing data on the CLL genetic background and landscape, including the role of many gene variants in disease initiation and progression in the context of telomere length, may be crucial for understanding CLL pathogenesis.30,31 This prompted us to investigate the role of MNS16A VNTR and selected SNPs located within the TERT gene in relation to telomere length and stage of disease in non-treated patients with CLL.
TERT SNPs were chosen using the SNP Function Prediction tool of the National Institute of Environmental Health Sciences website and other auxiliary databases (
 | Figure 1 Genomic structure of the human telomerase gene. Exons are marked in gray while intronic regions are in white. |
Materials and methods
Patients and controls
This retrospective study was conducted on a group of 68 treatment-naïve CLL patients (41 men and 27 women; aged 39–85 years, average 65.8 years) recruited at the Department of Haematology, Blood Neoplasms and Bone Marrow Transplantation, Wroclaw Medical University. Whole blood taken on EDTA from CLL patients and healthy individuals was used for genetic analysis and to assess telomere length.
Written informed consent was provided by the patients. The study was approved by the Wroclaw Medical University Ethics Committee and all the procedures were in accordance with the ethical standards of the Helsinki Declaration, as revised in 2013.
Stage of the disease was graded according to the Rai and Binet staging systems, based on clinical and laboratory parameters.30,31 Patients with CLL who were characterized with lower stages of disease (0–I in the Rai, and A in the Binet criteria) constituted approximately 70% of patients. The remaining patients were characterized with II–IV and B or C stage of disease at diagnosis. Patients’ characteristics are shown in Table 1. Genetic variants of the TERT gene and telomere length were assessed in the patients. In addition, blood donors served as controls for the polymorphism studies.
 | Table 1 TERT SNP genotype and allele distribution in CLL patients and controls |
Human cancer and leukemia cell lines
The following human cancer and leukemia cell lines (all commercially available) were used in the study: A549 (Sigma-Aldrich, Steinheim, Germany); HT-29, MV-4-11, HL-60, MDA-MB-231, Hs294T (American Type Culture Collection, Rockville, MD, USA); and KG1a, K562 (Leibniz-Institut DSMZ – German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany). They were maintained in liquid nitrogen at the Cell Culture Collection of the Hirszfeld Institute of Immunology and Experimental Therapy (Wroclaw, Poland). Cell line characteristics and culture conditions are presented in Table S1.
DNA isolation
DNA was isolated from 106 cultured cells of cancer and leukemia cell lines or from 5 mL of whole blood from CLL patients taken on EDTA using the Qiagen DNA Isolation Kit (Qiagen, Hilden, Germany) and following the recommendations of the manufacturer.
SNP genotyping of the TERT gene
The TERT polymorphic variants (rs2736100, rs2853690, rs33954691, rs35033501) in patients and controls were detected with the use of LightSNiP typing assays (TIB MOLBIOL, Berlin, Germany), employing real-time polymerase chain reaction (PCR) amplifications with melting curve analysis. The reactions were performed on a LightCycler 480 II Real-Time PCR system (Roche Diagnostics International, Rotkreuz, Switzerland), following the recommendations of the manufacturer.
VNTR MNS16A genotyping of the TERT gene
Presence of the MNS16A TERT gene polymorphism was assessed in all CLL patients and in healthy individuals. PCR was carried out in a 2720 Thermal Cycler instrument (Applied Biosystems, Foster City, CA, USA) using forward and reverse primer sequences (5ʹ-AGGATTCTGATCTCTGAAGGGTG-3ʹ and 5ʹ-TAMRA-TCTGCCTGAGGAAGGACGTATG-3ʹ) prepared by Genomed (Warsaw, Poland), based on Wang et al.24 The amplification procedure consisted of an initial denaturing step for 5 minutes at 95°C, followed by 35 cycles of: 30 seconds at 95°C, 45 seconds at 60°C and 1 minute at 72°C, as well as a final extension step for 10 minutes at 72°C. PCR products were diluted with formamide and GeneScan-500 ROX size standard (Applied Biosystems). Samples were denatured at 95°C for 5 minutes and quickly transferred to ice before analysis on the 3500 Genetic Analyzer (Applied Biosystems) with an eight-capillary system filled with the POP7 polymer. Alleles were identified with GeneMapper version 4.2 software (Applied Biosystems).
Telomere length analysis in CLL patients and human cancer cell lines
The average telomere length of target genomic DNA samples from human cell lines cultured in vitro and from whole blood of CLL patients was assessed by real-time quantitative polymerase chain reaction (qPCR) in a LightCycler480 II Real-Time PCR system (Roche Diagnostics International, Rotkreuz, Switzerland) using SYBR® Green assay kits (ScienCell’s Absolute Human Telomere Length Quantification qPCR Assay Kit [AHTLQ], Carlsbad, CA, USA), following the recommendations of the manufacturers. For each DNA sample, two consecutive reactions were performed: the first to amplify a single-copy reference (SCR) gene and the second for the telomere sequence. The SCR primer set recognizes and amplifies a 100 bp-long region on human chromosome 17 and serves as a reference for data normalization. Both PCRs were performed in a final volume of 20 μL using 1 μL of reference/genomic DNA samples from patients and controls (5 ng), 2 μL of primer stock solution (telomere or SCR), 10 μL of 2×qPCR FastStart Essential DNA Green Master Mix (Roche Diagnostics International) and 7 μL of nuclease-free water. The PCR conditions were as follows: 95°C for 10 minutes followed by 32 cycles of: 95°C for 20 seconds, 52°C for 20 seconds and 72°C for 45 seconds. All reactions were run in duplicate.
Statistical analysis
Genotype and allele frequencies were compared between the study groups by the chi-squared test with Yates' correction or Fisher’s exact test when necessary, using online tools (available online:
Results
TERT rs35033501 A variant as a potential genetic factor affecting CLL development
Minor allele frequencies in the healthy control group of the current study did not differ from those reported previously for Europeans, taken from the NCBI website (
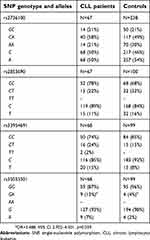
Comparison of the SNP genotypes and allele frequencies (rs2736100, rs2853690 and rs33954691) between our CLL patients and healthy subjects did not show significant differences, suggesting no association with disease predisposition. However, it was observed that CLL patients were more often characterized by the presence of the rs35033501 A allele than healthy individuals (9/68 vs 4/99, OR=3.488, 95% CI 2.702–4.501, p=0.039), implying that the rs35033501 SNP may affect disease susceptibility (Table 1).
Distribution of various MNS16A VNTR genotypes and alleles: association with CLL stage at diagnosis
Six different alleles and 11 genotypes of MNS16A were identified and classified as either long (L) or short (S) according to the length of PCR fragments.24,38 VNTR-364, VNTR-333 and VNTR-302 were marked as long (L) and VNTR-212, VNTR-243 and VNTR-274 as short (S).24 Based on this classification, various MNS16A genotypes were assigned to the three genotype groups: SS, SL or LL.38,39 Five different VNTRs of MNS16A were detected in our CLL patients (VNTR-243, VNTR-274, VNTR-302, VNTR-333 and VNTR-364) and four in healthy controls (VNTR-243, VNTR-274, VNTR-302, VNTR-333). Both groups harbored nine different genotypes. The MNS16A genotype and allele distributions in healthy controls are similar to other European populations, as reported by Andersson et al40 and Carpentier et al.41 The CLL patients and healthy subjects did not show significant differences in the MNS16A genotypes and allele frequencies. Genotype and allele distributions among CLL cases and healthy controls are presented in Table 2.
 | Table 2 TERT MNS16A genotype and allele distribution in CLL patients and controls |
Some differences were noticed when stage of the disease at diagnosis was considered. According to the Rai and Binet criteria,34,35 CLL patients older than 60 years were more frequently characterized by lower disease stage at diagnosis compared to patients aged 60 or younger (36/45 vs 9/23, p=0.001; 38/45 vs 12/23, p=0.008, for patients >60 years vs ≤60 years for the Rai and Binet staging systems, respectively) (Figure 2).
Moreover, in CLL patients who had a low disease stage at diagnosis (0–I and A), the MNS16A VNTR-243 (short allele) occurred more frequently than in patients with advanced stage (II–IV, B–C), according to the Rai (28/45 vs 7/23, p=0.020) and Binet systems (30/50 vs 5/18, p=0.028) (Figure 3). Thus, presence of the MNS16A VNTR-243 allele was more common in older patients with less advanced disease.
Relationships between VNTR MNS16A alleles and TERT SNP variants
The distribution of all genetic variants was compared between rs2736100, rs2853690, rs33954691 and rs35033501 polymorphisms and MNS16A tandem repeats. The TERT SNPs and MNS16A VNTR were not found to be in linkage disequilibrium (analyzed with Haploview 4.2 software:
Telomere length in CLL patients and in vitro cultured cell lines
Telomere length was analyzed in whole blood samples of 67 CLL patients at diagnosis (before treatment) and in eight cancer and leukemic cell lines cultured in vitro. The median of telomere length in patients was 5.71 kb. The median of telomere length in cell lines was even shorter and equaled 1.09 kb (Table S1). Both in CLL patients and in in vitro cultured cell lines, telomere length was shorter than the median telomere length of 7.54 kb and 7.26 kb reported for healthy donors by Jebaraj et al36 and Dos Santos et al,37 respectively.
No difference was observed between telomere length of female and male patients. Similarly, patients below and over 60 years of age did not show significant differences in telomere length. The latter observation is in line with the previous studies reporting a lack of correlation between telomere length and age of CLL patients.9,10,42
Relationship between telomere length and VNTR and SNP genetic variants in CLL patients
It appeared that telomeres were longer in CLL patients (N=44) with less advanced stage of disease (0–I) compared to CLL patients (N=23) with more advanced disease (II–IV stage), according to the Rai criteria (7.95 vs 5.99 kb, p=0.030). Moreover, among CLL patients carrying the TERT rs2736100 C allele, telomeres were longer in patients (N=40) in a less advanced stage of disease (Binet A) in comparison to those (N=13) at Binet stage B or C (7.50 vs 4.55 kb, p=0.048) (Figure 5A). A similar relationship was observed when the Rai criteria were considered. In patients in a less advanced stage (0–I) possessing the TERT rs2736100 C allele (N=36), significantly longer telomeres were detected than in those carrying the C allele but diagnosed with stages II–IV (N=17) (7.86 vs 4.48 kb, p=0.010) (Figure 5B).
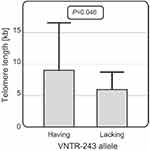
It was also observed that among CLL patients over 60 years of age, those carrying the VNTR-234 allele (N=27) had longer telomeres than patients lacking this allele (N=17) (with an average telomere length of 9.01 vs 6.03 kb, p=0.046) (Figure 6). For the other SNPs studied, no significant associations with telomere length were observed.
Discussion
Genetic variability within many genes can modulate telomere length and thus such genetic variants may constitute risk factors for the development of cancer and non-neoplastic diseases.43,44 Several studies suggest that genetically determined longer telomere length in peripheral leukocytes, also related with the TERT gene variants, could be associated with an increased risk of CLL.16 Also, a positive relationship between telomere length and multiple non-Hodgkin's lymphoma (NHL) subtypes, particularly for CLL/SLL (small lymphocytic lymphoma), was reported.15
The present retrospective study was conducted on a group of newly diagnosed untreated CLL patients. Assessment of telomere length, genetic factors related to TERT gene polymorphism, and identification of their relationships with susceptibility and clinical course of the disease were the key goals of this work. Therefore, four SNPs (rs2736100, rs2853690, rs33954691 and rs35033501) and the VNTR MNS16A gene polymorphism, all located within the TERT gene, were analyzed.
Results of a previous work by Ojha et al suggested some associations between CLL risk and the rs2736100 C allele that were not observed in the present study.16 Comparison of rs2736100, rs2853690, rs33954691, rs35033501 and VNTR MNS16A genotypes and allele frequencies between our CLL patients and healthy individuals did not indicate significant differences, except for the rs35033501 minor A variant. This variant was more frequently detected in CLL patients than in healthy controls, and was associated with a greater than three-fold increase in risk for CLL development. The rs35033501 polymorphism is a synonymous substitution that does not affect the amino acid sequence. However, such silent variants may potentially cause changes of splicing patterns.
Many recent studies have focused on the role of various factors affecting splicing in patients with CLL. The study by Palma et al aimed to characterize hTERT splice variants in CLL cells, as well as to examine the expression of hTERT splice variants and telomere length in relation to disease activity and clinical stage. The authors described two splicing sites that generate shorter transcripts and one full-length transcript that is translated into a functional protein. They observed that all transcripts were more frequently expressed in progressive than non-progressive patients and showed that average full-length transcript expression was 5.5-fold higher in immunoglobulin heavy chain variable region (IGHV)-unmutated CLL patients than in IGHV-mutated patients.44
A mutation within the splicing factor SF3B1 (splicing factor 3b subunit 1) gene in CLL was identified.45 SF3B1 encodes a protein involved in binding of the spliceosomal U2 snRNP to the branch point of the 3′ splicing site. Truncation of the protein by the introduction of premature stop codons is the most common outcome of splicing aberrations induced by SF3B1 mutations, affecting 90% of cases. In CLL, SF3B1 mutations are more frequent in later stages of the disease; they are associated with markers of poor clinical outcome and predict poor prognosis.45
In addition, genetic variability within other genes was reported to affect splicing in CLL patients. Puente et al identified novel recurrent mutations in non-coding regions, including the 39th region of notch receptor 1 (NOTCH1), which cause aberrant splicing events, increase NOTCH1 activity and result in a more aggressive disease.46
The aforementioned results show that various factors could affect splicing in patients with CLL, including genetic variability within non-coding regions also observed for the TERT rs35033501 SNP in the present study. Furthermore, some of these factors could be related to clinical parameters of the disease.
The polymorphic number of MNS16A tandem repeats was reported to be associated with the risk of several malignancies, including lung24,47 or colorectal and prostate cancer,38,39 malignant gliomas,40,41 as well as Alzheimer’s disease.48 So far, the MNS16A VNTR polymorphism of the hTERT catalytic subunit has been described in two lymphoproliferative disorders: first in our study on NHL,27 and later in diffuse large B-cell lymphoma (DLBCL).49 Our previous work identified some relationships between the VNTR-243 variant with more aggressive disease and with less favorable response to therapy.27 The study on DLBCL revealed that Egyptian carriers of the S allele or the SS genotype of MNS16A were at higher risk of disease development.49 Wang et al investigated promoter activity of MNS16A VNTRs in lung cancer cells and showed that the shorter VNTR (S) variant correlated with lower promoter activity, while the longer VNTR (L) was associated with increased risk of lung cancer.24 Hofer et al investigated promoter activity of all six known MNS16A VNTRs in different cell lines and showed the distribution of relative promoter activities of different MNS16A VNTRs determined by luciferase reporter assays. In all investigated cell lines, promoter activity of shorter constructs (also VNTRs-234) was higher than promoter activity of longer constructs, reflecting an indirect correlation between VNTR length and promoter activity.25 In addition, Zhang et al observed that carriers of the SL genotype had lower TERT expression compared to LL carriers when analyzing nasopharyngeal carcinoma tissue by immunohistochemical staining.26
The current results suggest a lack of association between the MNS16A TERT genetic polymorphism and predisposition to CLL. However, there were some interesting observations in regard to the MNS16A VNTR-243 short allele, which was more frequently detected in patients with less advanced disease (Rai 0–I and Binet A) than in patients with Rai II–IV and Binet B or C. These results are consistent with previous data showing a correlation between the presence of short MNS16A genotypes or alleles and advanced age at diagnosis of patients with prostate cancer or nasopharyngeal carcinoma and breast cancer.38,50 They imply that the presence of the MNS16A VNTR-243 short allele may affect the course of the disease. Indeed, in our previous study the presence of the MNS16A VNTR-243 short allele was found to play a role in progression and response to therapy of NHL patients.27
In various cancer scenarios, a dual role of telomere length has been observed. In general, in most patients who develop cancer the presence of long telomeres in tumor-initiating cells was detected. However, it is also well documented that critically short telomeres may lead to chromosomal instability, which can cause tumorigenesis. As for CLL, telomere dysfunction was observed in advanced stages of the disease, when the presence of critically short telomeres correlated with the occurrence of many genome rearrangements.11,12,51–53 Moreover, several studies showed that early-stage CLL patients exhibited extensive telomere erosion and fusion, indicating that telomere shortening and dysfunction can precede clinical progression.9,54 Also, no significant difference was observed between telomere length measured in tumor cells and in healthy cells of these CLL patients (Binet stage A and Rai stage 0). The study by Hoxha et al indicated the association between telomere status and genomic instability in CLL and found a significant correlation between telomere shortening and DNA hypomethylation in an early phase of disease.9 These results suggested that impairment of the telomere/telomerase system may represent an early event in CLL pathogenesis.9
We observed that CLL patients at a less advanced stage (Rai 0–I) at diagnosis had significantly longer telomeres than those with Rai stage II–IV, although their median telomere length equaled 5.71 kb and was shorter than median telomere length of 7.54 kb and 7.26 kb reported for healthy donors by Jebaraj et al36 and Dos Santos et al,37 respectively.
Moreover, significantly longer telomeres were observed among patients with less advanced disease (Rai 0–I or Binet A) possessing the TERT rs2736100 C allele, compared to patients with the TERT rs2736100 C allele, but exhibiting more advanced CLL stage (Rai III–IV or Binet B–C) (7.50 vs 4.55 kb and 7.86 vs 4.48 kb, for the Rai and Binet criteria, respectively). The observations above are in line with data from the Ojha et al study, which described the rs2736100 C allele as being associated with long leukocyte telomere length in a group of CLL patients.16
A common genetic variant in the TERT gene, rs2736100 C/A, is associated with both telomere length and risk in various diseases. This effect, however, was not directly visible in our CLL patients. A large meta-analysis performed by Snetselaar et al showed various effects of the TERT rs2736100 polymorphism in different disease association studies.55 Cancer cases were more often characterized by the presence of the TERT rs2736100 C allele, while for non-malignant diseases positive associations of the TERT rs2736100 A allele with disease risk were observed.55 Thus, the results of Snetselaar et al reflect a fundamentally different role of telomere biology in malignancies as opposed to non-malignant diseases, and illustrate the duality of telomere biology in different disease predisposition.55 In addition, the strength of the effect of rs2736100 polymorphism may vary between populations, as shown, for example, for Swedish and Chinese males with myeloproliferative neoplasms by Dahlström et al.56
In conclusion, the current results focusing on SNPs and the MNS16A polymorphism of the TERT gene and telomere length in CLL patients may add new valuable information to the knowledge regarding potential significance of telomere length/telomerase activity and expression in hematological malignancies. We showed that older CLL patients, who carried the MNS16A VNTR-243 short allele, were characterized by lower disease stage at diagnosis. That may be a result of a higher telomerase activity than that observed in patients with the long/long genotype and the long allele. Carriers of the long allele were previously shown to exhibit stronger TERT promoter activity and to carry the highest number of GATA-1 transcription factor binding sites, which leads to increased expression of antisense TERT mRNA and silencing of the sense telomerase transcript.
As SNPs may affect mRNA expression via splicing sites and/or miRNA binding site modifications, their detection within the TERT gene offers new prognostic opportunities. The current results also show an association between the rs35033501 A allele and disease susceptibility. Moreover, especially among patients possessing the TERT rs2736100 C allele, an association between telomere length and severity of the disease at diagnosis was observed.
Our results may help researchers to understand CLL development and identify new genetic biomarkers involved in this disease. The present study was particularly novel because it was the first to address the issue of TERT genetic variants (a VNTR and SNPs) in relation to telomere length in a hematological disorder. However, we are aware that our results should be further supported by functional tests of TERT expression in large groups of patients. Nevertheless, understanding the TERT polymorphism and expression in the context of CLL progression is an important step toward finding new ways to improve clinical care.
Abbreviations
CLL, chronic lymphocytic leukemia; DLBCL, diffuse large B-cell lymphoma; hTERT, human telomerase reverse transcriptase; IGHV, immunoglobulin heavy chain variable; L, long VNTR allele; qPCR, quantitative polymerase chain reaction; S, short VNTR allele; SCR, single-copy reference; SNP, single-nucleotide polymorphism; VNTR, variable number of tandem repeats.
Acknowledgments
The authors are grateful to Professor Joanna Wietrzyk and Dr Artur Anisiewicz from the Department of Experimental Oncology, Hirszfeld Institute of Immunology and Experimental Therapy, Polish Academy of Sciences, Wroclaw, Poland, for providing cancer cell lines. This work was supported by project TARGETTELO number STRATEGMED3/306853 from the National Centre for Research and Development, Warsaw, Poland.
Author contributions
All authors contributed toward data analysis, drafting and revising the paper, gave final approval of the version to be published and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Chiorazzi N, Rai KR, Ferrarini M. Chronic lymphocytic leukemia. N Engl J Med. 2005;352(8):804–815. doi:10.1056/NEJMoa040975
2. Rodríguez-Vicente AE, Díaz MG, Hernández-Rivas JM. Chronic lymphocytic leukemia: a clinical and molecular heterogenous disease. Cancer Genet. 2013;206(3):49–62. doi:10.1016/j.cancergen.2013.01.003
3. Friedman DR, Lucas JE, Weinberg JB. Clinical and biological relevance of genomic heterogeneity in chronic lymphocytic leukemia. PLoS One. 2013;8(2):e57356. doi:10.1371/journal.pone.0057356
4. Karakosta M, Delicha EM, Kouraklis G, Manola KN. Association of various risk factors with chronic lymphocytic leukemia and its cytogenetic characteristics. Arch Environ Occup Health. 2016;71(6):317–329. doi:10.1080/19338244.2015.1116429
5. Kasar S, Brown JR. Mutational landscape and underlying mutational processes in chronic lymphocytic leukemia. Mol Cell Oncol. 2016;3(4):e1157667. doi:10.1080/23723556.2016.1157667
6. Berndt SI, Camp NJ, Skibola CF, et al. Meta-analysis of genome-wide association studies discovers multiple loci for chronic lymphocytic leukemia. Nat Commun. 2016;9(7):10933. doi:10.1038/ncomms10933
7. Law PJ, Berndt SI, Speedy HE, et al. Genome-wide association analysis implicates dysregulation of immunity genes in chronic lymphocytic leukaemia. Nat Commun. 2017;6(8):14175. doi:10.1038/ncomms14175
8. Speedy HE, Di Bernardo MC, Sava GP, et al. A genome-wide association study identifies multiple susceptibility loci for chronic lymphocytic leukemia. Nat Genet. 2014;46(1):56–60. doi:10.1038/ng.2843
9. Hoxha M, Fabris S, Agnelli L, et al. Relevance of telomere/telomerase system impairment in early stage chronic lymphocytic leukemia. Genes Chromosomes Cancer. 2014;53(7):612–621. doi:10.1002/gcc.22171
10. Roos G, Kröber A, Grabowski P, et al. Short telomeres are associated with genetic complexity, high-risk genomic aberrations, and short survival in chronic lymphocytic leukemia. Blood. 2008;111(4):2246–2252. doi:10.1182/blood-2007-05-092759
11. Mansouri L, Grabowski P, Degerman S, et al. Short telomere length is associated with NOTCH1/SF3B1/TP53 aberrations and poor outcome in newly diagnosed chronic lymphocytic leukemia patients. Am J Hematol. 2013;88(8):647–651. doi:10.1002/ajh.23466
12. Rampazzo E, Bonaldi L, Trentin L, et al. Telomere length and telomerase levels delineate subgroups of B-cell chronic lymphocytic leukemia with different biological characteristics and clinical outcomes. Haematologica. 2012;97(1):56–63. doi:10.3324/haematol.2011.049874
13. Medves S, Auchter M, Chambeau L, et al. A high rate of telomeric sister chromatid exchange occurs in chronic lymphocytic leukaemia B-cells. Br J Haematol. 2016;174(1):57–70. doi:10.1111/bjh.13995
14. Poole JC, Andrews LG, Tollefsbol TO. Activity, function, and gene regulation of the catalytic subunit of telomerase (hTERT). Gene. 2001;269(1–2):1–12.
15. Machiela MJ, Lan Q, Slager SL, et al. Genetically predicted longer telomere length is associated with increased risk of B-cell lymphoma subtypes. Hum Mol Genet. 2016;25(8):1663–1676. doi:10.1093/hmg/ddw027
16. Ojha J, Codd V, Nelson CP, et al. ENGAGE consortium telomere group. genetic variation associated with longer telomere length increases risk of chronic lymphocytic leukemia. Cancer Epidemiol Biomarkers Prev. 2016;25(7):1043–1049. doi:10.1158/1055-9965.EPI-15-1329
17. Berndt SI, Skibola CF, Joseph V, et al. Genome-wide association study identifies multiple risk loci for chronic lymphocytic leukemia. Nat Genet. 2013;45(8):868–876. doi:10.1038/ng.2652
18. Mocellin S, Verdi D, Pooley KA, et al. Telomerase reverse transcriptase locus polymorphisms and cancer risk: a field synopsis and meta-analysis. J Natl Cancer Inst. 2012;104(11):840–854. doi:10.1093/jnci/djs222
19. Ropio J, Merlio JP, Soares P, Chevret E. Telomerase activation in hematological malignancies. Genes (Basel). 2016;7(9):
20. Zhdanov DD, Vasina DA, Grachev VA, et al. Alternative splicing of telomerase catalytic subunit hTERT generated by apoptotic endonuclease EndoG induces human CD4+ T cell death. Eur J Cell Biol. 2017;96(7):653–664. doi:10.1016/j.ejcb.2017.08.004
21. Zhdanov DD, Gladilina YA, Pokrovskaya MV, et al. Induction of alternative splicing and inhibition of activity of telomerase catalytic subunit by apoptotic endonuclease EndoG in human T, B, and NK cells. Bull Exp Biol Med. 2018;164(4):478–482. doi:10.1007/s10517-018-4016-y
22. Ghosh A, Saginc G, Leow SC, et al. Telomerase directly regulates NF-κB-dependent transcription. Nat Cell Biol. 2012;14(12):1270–1281. doi:10.1038/ncb2464
23. Heidenreich B, Kumar R. Altered TERT promoter and other genomic regulatory elements: occurrence and impact. Int J Cancer. 2017;141(5):867–876. doi:10.1002/ijc.30735
24. Wang L, Soria JC, Chang YS, et al. Association of a functional tandem repeats in the downstream of human telomerase gene and lung cancer. Oncogene. 2003;22(46):7123–7129. doi:10.1038/sj.onc.1206852
25. Hofer P, Zöchmeister C, Behm C, et al. MNS16A tandem repeat minisatellite of human telomerase gene: functional studies in colorectal, lung and prostate cancer. Oncotarget. 2017;8(17):28021–28027. doi:10.18632/oncotarget.15884
26. Zhang Y, Zhang H, Zhai Y, et al. A functional tandem-repeats polymorphism in the downstream of TERT is associated with the risk of nasopharyngeal carcinoma in Chinese population. BMC Med. 2011;9:1–9. doi:10.1186/1741-7015-9-106
27. Wysoczanska B, Wrobel T, Dobrzynska O, Mazur G, Bogunia-Kubik K. Role of the functional MNS16A VNTR- 243 variant of the human telomerase reverse transcriptase gene in progression and response to therapy of patients with non-Hodgkin’s B-cell lymphomas. Int J Immunogenet. 2015;42(2):100–105. doi:10.1111/iji.2015.42.issue-2
28. Concetti F, Lucarini N, Carpi FM, et al. The functional VNTR MNS16A of the TERT gene is associated with human longevity in a population of Central Italy. Exp Gerontol. 2013;48(6):587–592. doi:10.1016/j.exger.2013.03.009
29. Chen P, Zou P, Yan Q, et al. The TERT MNS16A polymorphism contributes to cancer susceptibility: meta-analysis of the current studies. Gene. 2013;519(2):266–270. doi:10.1016/j.gene.2013.02.018
30. Xia X, Rui R, Quan S, et al. MNS16A tandem repeats minisatellite of human telomerase gene and cancer risk: a meta-analysis. PLoS One. 2013;8(8):e73367. doi:10.1371/journal.pone.0073367
31. Fabbri G, Dalla-Favera R. The molecular pathogenesis of chronic lymphocytic leukaemia. Nat Rev Cancer. 2016;16(3):145–162. doi:10.1038/nrc.2016.8
32. Codd V, Nelson CP, Albrecht E, et al. Identification of seven loci affecting mean telomere length and their association with disease. Nat Genet. 2013;45(4):422–427. doi:10.1038/ng.2528
33. Guieze R, Wu CJ. Genomic and epigenomic heterogeneity in chronic lymphocytic leukemia. Blood. 2015;126(4):445–453. doi:10.1182/blood-2015-02-585042
34. Rai KR, Sawitsky A, Cronkite EP, et al. Clinical staging of chronic lymphocytic leukemia. Blood. 1975;46(2):219–234.
35. Binet JL, Auquier A, Dighiero G, et al. A new prognostic classification of chronic lymphocytic leukemia derived from a multivariate survival analysis. Cancer. 1981;48(1):198–204.
36. Jebaraj BM, Kienle D, Lechel A, et al. Telomere length in mantle cell lymphoma. Blood. 2013;121(7):1184–1187. doi:10.1182/blood-2012-08-452649
37. Dos Santos P, Panero J, Palau Nagore V, et al. Telomere shortening associated with increased genomic complexity in chronic lymphocytic leukemia. Tumour Biol. 2015;36(11):8317–8324. doi:10.1007/s13277-015-3556-2
38. Hofer P, Baierl A, Feik E, et al. MNS16A tandem repeats minisatellite of human telomerase gene: a risk factor for colorectal cancer. Carcinogenesis. 2011;32(6):866–871. doi:10.1093/carcin/bgr053
39. Hofer P, Zerelles J, Baierl A, et al. MNS16A tandem repeat minisatellite of human telomerase gene and prostate cancer susceptibility. Mutagenesis. 2013;28(3):301–306. doi:10.1093/mutage/get003
40. Andersson U, Osterman P, Sjöström S, et al. MNS16A minisatellite genotypes in relation to risk of glioma and meningioma and to glioblastoma outcome. Int J Cancer. 2009;125(4):968–972. doi:10.1002/ijc.24363
41. Carpentier C, Lejeune J, Gros F, et al. Association of telomerase gene hTERT polymorphism and malignant gliomas. J Neurooncol. 2007;84(3):249–253. doi:10.1007/s11060-007-9378-3
42. Steinbrecher D, Jebaraj BMC, Schneider C, et al. Telomere length in poor-risk chronic lymphocytic leukemia: associations with disease characteristics and outcome. Leuk Lymphoma. 2018;59(7):1614–1623. doi:10.1080/10428194.2017.1390236
43. Haycock PC, Burgess S, Nounu A, et al. Association between telomere length and risk of cancer and non-neoplastic diseases: a mendelian randomization study. Telomeres mendelian randomization collaboration. JAMA Oncol. 2017;3(5):636–651. doi:10.1001/jamaoncol.2016.5945
44. Palma M, Parker A, Hojjat-Farsangi M, et al. Telomere length and expression of human telomerase reverse transcriptase splice variants in chronic lymphocytic leukemia. Exp Hematol. 2013;41(7):615–626. doi:10.1016/j.exphem.2013.03.008
45. Anczuków O, Krainer AR. Splicing-factor alterations in cancers. Rna. 2016;22(9):1285–1301. doi:10.1261/rna.059303.116
46. Puente XS, Beà S, Valdés-Mas R, et al. Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nature. 2015;526(7574):519–524. doi:10.1038/nature14666
47. Jin G, Yoo SS, Cho S, et al. Dual roles of a variable number of tandem repeat polymorphism in the TERT gene in lung cancer. Cancer Sci. 2011;102(1):144–149. doi:10.1111/j.1349-7006.2010.01782.x
48. Scarabino D, Broggio E, Gambina G, et al. Common variants of human TERT and TERC genes and susceptibility to sporadic Alzheimers disease. Exp Gerontol. 2017;88:19–24. doi:10.1016/j.exger.2016.12.017
49. Essa ES, Alagizy HA. Association of MNS16A VNTR and hTERT rs2736098: G>A polymorphisms with susceptibility to diffuse large B-cell lymphoma. Tumori. 2018;104(3):165–171. doi:10.5301/tj.5000653
50. Zagouri F, Sergentanis TN, Gazouli M, et al. HTERT MNS16A polymorphism in breast cancer: a case–control study. Mol Biol Rep. 2012;39(12):10859–10863.
51. Lin TT, Letsolo BT, Jones RE, et al. Telomere dysfunction and fusion during the progression of chronic lymphocytic leukemia: evidence for a telomere crisis. Blood. 2010;116(111):1899–1907. doi:10.1182/blood-2010-02-272104
52. Sellmann L, de Beer D, Bartels M, et al. Telomeres and prognosis in patients with chronic lymphocytic leukaemia. Int J Hematol. 2011;93(1):74–82. doi:10.1007/s12185-010-0750-2
53. Sellmann L, Scholtysik R, de Beer D, et al. Shorter telomeres correlate with an increase in the number of uniparental disomies in patients with chronic lymphocytic leukemia. Leuk Lymphoma. 2016;57(3):590–595. doi:10.3109/10428194.2015.1076929
54. Lin TT, Norris K, Heppel NH, et al. Telomere dysfunction accurately predicts clinical outcome in chronic lymphocytic leukaemia, even in patients with early stage disease. Br J Haematol. 2014;167(2):214–223. doi:10.1111/bjh.13023
55. Snetselaar R, van Oosterhout MFM, Grutters JC, van Moorsel CHM. Telomerase reverse transcriptase polymorphism rs2736100: a balancing act between cancer and non-cancer disease, a meta-analysis. Front Med. (Lausanne). 2018;5:41. doi:10.3389/fmed.2018.00041
56. Dahlström J, Liu T, Yuan X, et al. TERT rs2736100 genotypes are associated with differential risk of myeloproliferative neoplasms in Swedish and Chinese male patient populations. Ann Hematol. 2016;95(11):1825–1832. doi:10.1007/s00277-016-2787-7
Supplementary material
 | Table S1 Telomere length in cancer and leukemia cell lines, cell line characteristics and culture conditions |
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.