Back to Journals » OncoTargets and Therapy » Volume 8
Validation of a quantitative 12-multigene expression assay (Oncotype DX® Colon Cancer Assay) in Korean patients with stage II colon cancer: implication of ethnic differences contributing to differences in gene expression
Authors Jeong DH, Kim WR, Min BS, Kim Y, Song MK, Kim NK
Received 1 September 2015
Accepted for publication 10 November 2015
Published 17 December 2015 Volume 2015:8 Pages 3817—3825
DOI https://doi.org/10.2147/OTT.S95543
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Faris Farassati
Duck Hyoun Jeong,1 Woo Ram Kim,1 Byung Soh Min,1 Young Wan Kim,2 Mi Kyung Song,3 Nam Kyu Kim1
1Department of Surgery, Yonsei University College of Medicine, Seoul, 2Department of Surgery, Wonju College of Medicine, Wonju, 3Department of Research Affairs, Biostatistics Collaboration Unit, Yonsei University College of Medicine, Seoul, Korea
Purpose: To evaluate the Recurrence Score® of the quantitative 12-multigene expression assay and to determine risk groups based on the continuous Recurrence Score® in Korean patients.
Method: A total of 95 patients with pathological T3N0 tumors and mismatch repair-proficient tumors were enrolled. The Recurrence Score® was used to classify risk groups (low risk, <30; intermediate risk, 30–40; high risk, ≥41).
Results: Fifty-four patients (56.8%) were aged over 70 years. There were 49 men (51.6%) and 56 cases of right-sided colon cancer (58.9%). Eight cases (8.4%) had well-differentiated tumors, and 86 cases (90.5%) showed moderate differentiation. Only one case (1.1%) had a poorly differentiated tumor. Three patients (3.2%) had lymphovascular invasion. Sixty-one patients were identified as low risk (64.2%) and 34 patients as intermediate risk (35.8%). There were no high-risk patients. Although not significant, the 3-year recurrence risk increased with the Recurrence Score®.
Conclusion: Distribution patterns of risk groups based on the Recurrence Score®, particularly the absence of a high-risk group, were different from the prior validation studies. These findings suggest that ethnic differences between Koreans and Western patients are potential contributing factors for different gene expressions in the quantitative 12-multigene expression assay.
Keywords: colonic neoplasms, gene expression, adjuvant chemotherapy, ethnic groups
Introduction
Colorectal cancer (CRC) is the third most common malignancy in the world.1 In Asia, including Korea, CRC is currently the fourth leading cause of mortality by cancer, and its prevalence is increasing.2 After curative-intent resection of colon cancer, adjuvant therapy is performed to eliminate any potential source of disease recurrence. The benefits of adjuvant chemotherapy have been widely proven for stage III colon cancer; thus, it has become a standard treatment for such cases. However, the role of adjuvant chemotherapy remains less clear in patients with stage II colon cancer.3–5
Outcomes of stage II colon cancer vary;6,7 accordingly, risk stratification within stage II disease has been performed using clinical and pathologic variables. The current National Comprehensive Cancer Network guidelines recommend adjuvant chemotherapy for stage II colon cancer when patients have a pathological (p)T4 lesion, intestinal perforation or obstruction, lymphovascular invasion, perineural invasion, poorly differentiated histology, or fewer than 12 lymph nodes examined.8 However, there are no convincing data to predict the recurrence risk accurately for stage II disease. Thus, it is necessary to identify more effective predictors that can be used in addition to the traditional clinicopathologic parameters to assist in recurrence risk stratification and treatment decision-making for patients with stage II colon cancer.
Multiple-gene analysis can provide more reliable insight into tumor biology than single-gene analysis, and it can also yield more robust information regarding each prognosis, diagnosis, and response to treatment. In recent years, several groups have developed multigene panel assays to determine the prognosis of patients with stage II colon cancer.9–11 One of these assays is the Oncotype DX® Colon Cancer Assay (Genomic Health, Redwood City, CA, USA), which utilizes a quantitative reverse transcription-polymerase chain reaction (RT-PCR)-based panel test using 12 genes. This panel was validated as a significant predictor for recurrence in stage II colon cancer.12–14
Most validation studies have been performed in Western countries;12–14 however, Ollberding et al15 suggested that ethnic difference may contribute to CRC risk through genetic susceptibility in their multiethnic cohort study. Thus, we postulated that gene expression patterns of the quantitative 12-multigene expression assay could differ between Western and Korean patients. The purpose of this pilot study was to investigate 1) the Recurrence Score® of the quantitative 12-multigene expression assay in Korean patients with stage II colon cancer and 2) the association between recurrence risk and the Recurrence Score® of the quantitative 12-multigene expression assay.
Methods
Patients and materials
This study included a retrospective study (n=56) and a prospective study (n=39). Eligibility criteria for the retrospective study was as follows: Korean patients with biopsy-proven adenocarcinoma derived from the colon who underwent curative resection for colon cancer from January 2008 to December 2010; a postoperative pathologic report of American Joint Committee on Cancer stage II colon cancer with mismatch repair (MMR) gene-proficiency (low or stable microsatellite instability); no adjuvant chemotherapy; and available freshly frozen tumor tissues. Exclusion criteria were high-risk features, including the presence of a pT4 lesion, MMR gene-deficiency (high microsatellite instability),16 and a lack of freshly frozen tumor tissue. We obtained freshly frozen tumor tissue samples and complete medical records of 56 patients who satisfied the eligibility criteria. Age, sex, tumor location, and evidence of bowel obstruction or perforation were obtained from clinical records. Events of recurrence were included as local and systemic recurrences. Local recurrence was defined as tumor regrowth in the vicinity of the primary tumor site, and systemic recurrence was defined as tumor recurrence in the distant organs. We used all fresh frozen samples, and this methodology differed from that used in the original study, which used fixed paraffin-embedded tissue samples. Unfortunately, we did not compare RNA quality between paraffin-embedded tissue and fresh frozen tissue samples. However, it has been reported that fresh frozen tissue preserves histologic cellular architecture and generates good-quality RNA.17
The prospective study enrolled 39 patients from January to December 2012 using the same eligibility criteria as were used for retrospective enrollment. Patients who provided written informed consent were enrolled, and this study was approved by the institutional review board of Yonsei University Health System (number 4-2011-0888). The tumor tissues were stored in the Severance Hospital Gene Bank, which has been in operation since 2005. Tumor samples were obtained within 1 hour after surgical resection, transferred to the Gene Bank immediately, and frozen with liquid nitrogen. Specimens were stored at −80°C until further processing.
Recurrence Score®
The 12-multigene Recurrence Score® was calculated using the prespecified genes and algorithm that were previously validated12 and recognized cutoff points for low-, intermediate-, and high-recurrence risk groups (Recurrence Scores of <30, 30–40, and ≥41, respectively).
Endpoints
The primary endpoint was to evaluate the Recurrence Score® of the quantitative 12-multigene expression assay and to determine risk groups based on a continuous Recurrence Score® in Korean patients with stage II colon cancer. The secondary end point was to evaluate the relationship between the continuous Recurrence Score® and the 3-year recurrence risk.
Histopathologic data
Tumor grade and type were centrally assessed according to the College of American Pathologists Consensus Statement by an academic surgical pathologist with subspecialty expertise in gastrointestinal pathology.18 Data for pathologic T stage, number of nodes examined, lymphovascular invasion, perineural invasion, and histologic grade were recorded. MMR gene status was assessed in accordance with the consensus definitions of the National Cancer Institute, as previously described.19
Gene expression analysis
RNA extraction
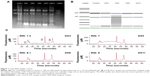
Total RNA extraction was performed using TRIzol® reagent according to the manufacturer’s instructions (Life Technologies, Carlsbad, CA, USA). After RNA isolation, total RNA was checked for the quantity, purity, and integrity of the 18S and 28S ribosomal bands. Total RNA concentration was determined using a Nanodrop spectrophotometer ND-1000 (Thermo Fisher Scientific, Waltham, MA, USA). For each sample quantity tested, RNA degradation was assessed using the Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA) (Figure 1).
Reverse transcription
The RT reaction was carried out with 1 μg of total RNA using the QuantiTect® Reverse Transcriptase Kit (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions.
Quantitative PCR (qPCR) and gene-expression analysis
cDNA for target genes (BGN, MYC, FAP, GADD45B, INHBA, MK167, MYBL2, UBB) and reference genes (ATP5E, GPX1, PGK1, UBB, and VDAC2) was distributed into 96-well plates and PCR forward and reverse primer and TaqMan probes were added. The TaqMan® probes and primers were designed as previously described.20 The primer and probe sequences used are presented in Table 1. Oligonucleotides were purchased from Cosmogenetech Co., Ltd. (Seoul, Korea). In this study, TaqMan probes had a fluorescein (6-FAM®) 5′-reporter and a Black Hole Quencher®-1 (BHQ-1®) 3′-quencher. qPCR was performed using a fluorescence-based real-time detection system (ABI PRISM 7000 sequence detection system; Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s instructions. Reactions were performed in a 25 μL volume with cDNA equivalent to 1 μg total RNA. The final primer and probe concentrations were 0.9 μmol/L (primers) and 0.25 μmol/L (probes). PCR cycling conditions were 95°C for one cycle of 10 minutes, followed by 43 cycles of 20 seconds at 95°C then 45 seconds at 60°C. Each gene assay was run in triplicate wells.
Reference gene normalization
Normalization was performed using the average expression of the five reference genes (ATP5E, GPX1, PGK1, UBB, and VDAC2). The mean cycle threshold (Ct) value for each gene was subtracted from the mean Ct for the five reference genes.20
Statistical analysis
All data are described as frequency and percentage. Differences among groups were analyzed with the chi-square test or Fisher’s exact test. Univariate and multivariate Cox regression analyses were used to estimate the hazard ratio (HR) and 95% confidence interval (CI). A rug plot was displayed to confirm the relationship between observed and predicted risk. All tests were two-sided, and a P-value of 0.05 was considered to be statistically significant. Statistical analyses were conducted using R project for Statistical Computing, Version 2.12.0 (R Development Core Team, Vienna, Austria) and SPSS software version 20.0 for Windows (SPSS Corp., Chicago, IL, USA).
Results
Demographics
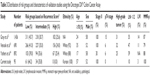
Fifty-four patients (56.8%) were aged over 70 years and the mean age was 69.6 years (range, 43–85). The patient group consisted of 49 (51.6%) men and 46 (48.4%) women. Fifty-six carcinomas (58.9%) were located in the right-sided colon, and 39 (41.1%) were in the left-sided colon. Eight cases (8.4%) had well-differentiated tumors, and 86 cases (90.5%) showed moderate differentiation. Only one case (1.1%) had a poorly differentiated tumor. Three patients (3.2%) had lymphovascular invasion (+) tumors and two patients (2.1%) had perineural invasion (+) tumors.
There were no differences in demographics between the retrospective and prospective cohorts except for the number of lymph nodes examined. In the retrospective cohort, nine patients (9.5%) had fewer than 12 harvested lymph nodes in the specimen; however, all patients had 12 or more nodes in the prospective cohort. Detailed demographics are summarized in Table 2.
Distribution of risk groups based on Recurrence Score®
The 12-multigene Recurrence Score® ranged from 0 to 39, with a median score of 28 (interquartile range, 20.3–31.8) and a mean of 23.3 (±11.4 standard deviation). Risk groups were classified based on the calculated Recurrence Score® (low risk, <30; intermediate risk, 30–40; high risk, ≥41). Sixty-one patients were identified as low risk (64.2%) and 34 as intermediate risk (35.8%). There were no high-risk patients (Table 3).
Relationship between Recurrence Score® and recurrence risk
To evaluate the relationship between the continuous Recurrence Score® and the 3-year recurrence risk, we performed subgroup analyses in the retrospective cohort. The median follow-up time was 31 months. During the follow-up period, four patients (4.2%) developed recurrence. Three cases were in the low-risk group and one was in the intermediate-risk group (Table 4). In the primary analysis based on the Cox model, although not significant, the 3-year recurrence risk increased with the Recurrence Score® (HR per interquartile range, 0.52; 95% CI, 0.19–1.39; P=0.58; Figure 2).
  | Table 4 Four patients with recurrence |
  | Figure 2 Relationship between the continuous Recurrence Score® and 3-year recurrence risk. |
Conventional clinical and pathologic factors including age, sex, tumor location, tumor grade, and number of lymph nodes did not show any significant associations with recurrence-free survival in the univariate and multivariate analyses using the Cox proportional hazard model. Similarly, risk assessment based on the National Comprehensive Cancer Network guidelines was not a significant risk factor for recurrence-free survival, with a HR of 1.41 (95% CI, 0.15–13.58; P=0.767; Table 5).
Discussion
We hypothesized that gene expression patterns of Korean patients may be different from those of Western patients. Thus, we determined gene expression using the quantitative 12-multigene expression assay, Recurrence Scores®, and risk groups (low, intermediate, or high risk). The distribution of risk groups based on Recurrence Score® showed that majority of patients (n=61, 64.2%) were in the low-risk group and the remaining were in the intermediate-risk group (35.8%). There were no high-risk patients. The distribution pattern of risk groups, particularly the absence of a high-risk group, was quite different from the published validation studies.12–14 These results suggest that ethnic differences between Korean and Western patients may play a role in the observed differences in gene expression from the quantitative 12-multigene expression assay. However, we acknowledge that other underlying factors that were not considered in this study might have influenced differences in gene expression. In addition, the Recurrence Score® predicted a recurrence risk in the previous validation studies,12–14 and we also found that, although not significant, Recurrence Score® was associated with an increased 3-year recurrence risk. The nonsignificant result could be attributed to the small study sample.
Detailed clinical and pathologic variables of major validation studies are summarized in Table 3. Upon a review of the literature, the distribution pattern of risk groups (notably the absence of high-risk patients in this study) differed from major validation studies, such as the QUASAR (Quick And Simple And Reliable) trial,12 CALGB (Cancer And Leukemia Group B) 9581 trial,13 and NSABP (National Surgical Adjuvant Breast and Bowel Project)-C07 study.14 The proportion of low-risk group patients was 39.0%–43.7% in the prior studies12–14 yet was 64.2% in this study. Additionally, the proportion of Caucasian patients (White race/ethnicity) was predominant (88%–92%) in the previous studies.13,14 Our results suggest that the genetic characteristics of CRC in Asian patients, particularly Koreans, are different from Western patients based on results of the quantitative 12-multigene expression assay for colon cancer.
In previous studies, the proportion of patients older than 70 years was 17%–35%, and the rate of less than 12 lymph nodes ranged from 35% to 47%;12–14 however, our study yielded corresponding results of 57% and 10%. Interestingly, high-grade tumors were much more uncommon (1%) in this study than in prior studies (25%–32%).12–14 Gray et al12 observed that high-grade tumors were more frequent in the high-risk group than in the low-risk group (44% vs 23%). The authors suggested that patients with higher Recurrence Score® were more likely to have T4 disease, MMR-deficient tumors, and high-grade tumors. It is possible that, by excluding those with T4 disease and those with MMR-deficient tumors, our sampling process excluded patients with high-grade tumors. The lower Recurrence Scores® in this study population might be attributable to the limited inclusion criteria. Thus, if we had included more high-grade tumors, the proportion of tumors in the high-risk group would likely have increased. However, it is possible that the absence or relatively low proportion of high-risk patients could be distinctive genetic characteristics of Korean patients found using the quantitative 12-multigene expression assay. Thus, the validation results of Western patients need to be cautiously applied to other ethnicities, including Korean patients.
In this study, only pT3N0 and MMR-proficient tumors were included in order to maintain a homogenous study population. MMR gene-deficiency, also called high microsatellite instability, is associated with a lower recurrence risk in stage II colon cancer and is unlikely to benefit from fluorouracil-based adjuvant chemotherapy.19,21,22 Ethnic differences of MMR-gene status have also been shown. Hong et al19 reported that rates of MMR gene-deficient tumors were ~9% in Korean CRC patients and ~18% in Western patients.23,24 These differences in MMR gene status suggest that ethnic differences are associated with gene expression. Our finding based on results from the quantitative 12-multigene expression assay also supports the hypothesis that ethnic difference is associated with gene expression difference.
Three validation studies, the QUASAR trial,12 CALGB 9581 trial,13 and NSABP-C07 study,14 showed that Recurrence Score® was significantly associated with recurrence risk in stage II12,13 or stage II and III patients.14 The prognostic significance of the 12-multigene expression assay has also been validated in rectal cancer patients. Reimers et al25 reported that Recurrence Score® was a significant predictor for recurrence risk and cancer-specific survivals in stage II and III rectal cancer patients. In this study, we also observed a similar pattern in that the 3-year recurrence risk increased with Recurrence Score®, although this association was not significant. Our study had only four events of recurrences; thus, if there were more recurrence events in our study population, we could expect a significant relationship between Recurrence Score® and recurrence risk.
The main limitations of this study were the small study population and the fact that there were only four recurrence events; thus, it was difficult to obtain meaningful results from the multivariate risk factor analysis for recurrence-free survival. In addition, this study did not include patients with rectal cancer, which has previously been associated with more frequent recurrences compared with colon cancer.26 However, it was the first study to investigate gene expression patterns based on the quantitative 12-multigene expression assay in Korean patients with colon cancer. In addition, our study population was relatively homogenous in that it included only T3N0 tumors and MMR-proficient tumors.
In summary, distribution patterns of risk groups based on Recurrence Score®, especially absence of a high-risk group, were different from the results of prior validation studies.12–14 These findings suggest that ethnic differences between Korean and Western patients are potential contributing factors of differences in gene expression in the quantitative 12-multigene expression assay. In addition, although not significant, Recurrence Score® was associated with an increased 3-year recurrence risk. Based on our findings, recurrence risk stratification based on the quantitative 12-multigene expression assay can be applied to Korean patients with stage II and MMR gene-proficient tumors. However, the results of the assay should be interpreted cautiously in Korean patients, as unidentified ethnic differences may exist in the gene expression pattern. Moreover, there might have been other factors involved in differences in gene expression in the quantitative 12-multigene expression assay that were not addressed in this study. We did not evaluate another cohort of a different ethnicity. Accordingly, it should be determined in future research whether ethnicity is the main reason for the differences in Recurrence Score® in other ethnic groups. In addition, definite conclusions could not be drawn in this study due to the limited sample size. In future perspectives, it would be useful to identify single-nucleotide polymorphisms affecting gene expression levels of genes included in the quantitative 12-multigene expression assay in a Korean population, and more genetic studies should be performed in order to validate exact ethnic differences in gene expression.
Acknowledgments
This study was supported by a grant from the Korea Healthcare Technology R&D Project, Ministry of Health and Welfare, Republic of Korea (A102065). The funding source had no involvement in the study. This paper was presented in part at the 2014 American Society of Colon & Rectal Surgeons Annual Meeting, May 17–21, 2014, Hollywood, FL, USA.
Disclosure
The authors report no conflicts of interest in this work.
References
Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63(1):11–30. | ||
Park HC, Shin A, Kim BW, et al. Data on the characteristics and the survival of Korean patients with colorectal cancer from the Korea central cancer registry. Ann Coloproctol. 2013;29(4):144–149. | ||
Gray R, Barnwell J, McConkey C, Hills RK, Williams NS, Kerr DJ. Adjuvant chemotherapy versus observation in patients with colorectal cancer: a randomised study. Lancet. 2007;370(9604):2020–2029. | ||
Moertel CG, Fleming TR, Macdonald JS, et al. Intergroup study of fluorouracil plus levamisole as adjuvant therapy for stage II/Dukes’ B2 colon cancer. J Clin Oncol. 1995;13(12):2936–2943. | ||
Schippinger W, Samonigg H, Schaberl-Moser R, et al. A prospective randomised phase III trial of adjuvant chemotherapy with 5-fluorouracil and leucovorin in patients with stage II colon cancer. Br J Cancer. 2007;97(8):1021–1027. | ||
Figueredo A, Charette ML, Maroun J, Brouwers MC, Zuraw L. Adjuvant therapy for stage II colon cancer: a systematic review from the Cancer Care Ontario Program in evidence-based care’s gastrointestinal cancer disease site group. J Clin Oncol. 2004;22(16):3395–3407. | ||
Gill S, Loprinzi CL, Sargent DJ, et al. Pooled analysis of fluorouracil-based adjuvant therapy for stage II and III colon cancer: who benefits and by how much? J Clin Oncol. 2004;22(10):1797–1806. | ||
National Comprehensive Cancer Network. National comprehensive cancer network guidelines, Colon cancer (Version 2.2015). Available from: http://www.nccn.org/professionals/physician_gls/pdf/colon.pdf. Accessed May 1, 2015. | ||
O’Connell MJ, Lavery I, Yothers G, et al. Relationship between tumor gene expression and recurrence in four independent studies of patients with stage II/III colon cancer treated with surgery alone or surgery plus adjuvant fluorouracil plus leucovorin. J Clin Oncol. 2010;28(25):3937–3944. | ||
Agesen TH, Sveen A, Merok MA, et al. ColoGuideEx: a robust gene classifier specific for stage II colorectal cancer prognosis. Gut. 2012;61(11):1560–1567. | ||
Maak M, Simon I, Nitsche U, et al. Independent validation of a prognostic genomic signature (ColoPrint) for patients with stage II colon cancer. Ann Surg. 2013;257(6):1053–1058. | ||
Gray RG, Quirke P, Handley K, et al. Validation study of a quantitative multigene reverse transcriptase-polymerase chain reaction assay for assessment of recurrence risk in patients with stage II colon cancer. J Clin Oncol. 2011;29(35):4611–4619. | ||
Venook AP, Niedzwiecki D, Lopatin M, et al. Biologic determinants of tumor recurrence in stage II colon cancer: validation study of the 12-gene recurrence score in cancer and leukemia group B (CALGB) 9581. J Clin Oncol. 2013;31(14):1775–1781. | ||
Yothers G, O’Connell MJ, Lee M, et al. Validation of the 12-gene colon cancer recurrence score in NSABP C-07 as a predictor of recurrence in patients with stage II and III colon cancer treated with fluorouracil and leucovorin (FU/LV) and FU/LV plus oxaliplatin. J Clin Oncol. 2013; 31(36):4512–4519. | ||
Ollberding NJ, Nomura AM, Wilkens LR, Henderson BE, Kolonel LN. Racial/ethnic differences in colorectal cancer risk: the multiethnic cohort study. Int J Cancer. 2011;129(8):1899–1906. | ||
Kim NK, Park JK, Shin E, Kim YW. The combination of nuclear factor kappa B, cyclo-oxygenase-2 and vascular endothelial growth factor expression predicts poor prognosis in stage II and III colorectal cancer. Anticancer Res. 2014;34(11):6451–6457. | ||
Micke P, Ohshima M, Tahmasebpoor S, et al. Biobanking of fresh frozen tissue: RNA is stable in nonfixed surgical specimens. Lab Invest. 2006;86(2):202–211. | ||
Compton CC, Fielding LP, Burgart LJ, et al. Prognostic factors in colorectal cancer. College of American Pathologists Consensus Statement 1999. Arch Pathol Lab Med. 2000;124(7):979–994. | ||
Hong SP, Min BS, Kim TI, et al. The differential impact of microsatellite instability as a marker of prognosis and tumour response between colon cancer and rectal cancer. Eur J Cancer. 2012;48(8):1235–1243. | ||
Clark-Langone KM, Sangli C, Krishnakumar J, Watson D. Translating tumor biology into personalized treatment planning: analytical performance characteristics of the Oncotype DX Colon Cancer Assay. BMC Cancer. 2010;10:691. | ||
Kim H, Nam SW, Rhee H, et al. Different gene expression profiles between microsatellite instability-high and microsatellite stable colorectal carcinomas. Oncogene. 2004;23(37):6218–6225. | ||
Pino MS, Chung DC. Microsatellite instability in the management of colorectal cancer. Expert Rev Gastroenterol Hepatol. 2011;5(3):385–399. | ||
Kim GP, Colangelo LH, Wieand HS, et al. Prognostic and predictive roles of high-degree microsatellite instability in colon cancer: a National Cancer Institute-National Surgical Adjuvant Breast and Bowel Project Collaborative Study. J Clin Oncol. 2007;25(7):767–772. | ||
Ribic CM, Sargent DJ, Moore MJ, et al. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N Engl J Med. 2003;349(3):247–257. | ||
Reimers MS, Kuppen PJ, Lee M, et al. Validation of the 12-gene colon cancer recurrence score as a predictor of recurrence risk in stage II and III rectal cancer patients. J Natl Cancer Inst. 2014;106(11):1–8. | ||
Renouf DJ, Woods R, Speers C, et al. Improvements in 5-year outcomes of stage II/III rectal cancer relative to colon cancer. Am J Clin Oncol. 2013;36(6):558–564. |
 © 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.