Back to Journals » International Journal of Women's Health » Volume 7
Utility of proteomics in obstetric disorders: a review
Authors Hernández-Núñez J, Valdés-Yong M
Received 19 December 2014
Accepted for publication 5 March 2015
Published 13 April 2015 Volume 2015:7 Pages 385—391
DOI https://doi.org/10.2147/IJWH.S79577
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Elie Al-Chaer
Jónathan Hernández-Núñez,1 Magel Valdés-Yong2
1Department of Obstetrics and Gynecology, Hospital Alberto Fernández-Valdés, Santa Cruz del Norte, Mayabeque, 2Department of Obstetrics and Gynecology, Hospital Luis Díaz Soto, Habana del Este, La Habana, Cuba
Abstract: The study of proteomics could explain many aspects of obstetric disorders. We undertook this review with the aim of assessing the utility of proteomics in the specialty of obstetrics. We searched the electronic databases of MEDLINE, EBSCOhost, BVS Bireme, and SciELO, using various search terms with the assistance of a librarian. We considered cohort studies, case-control studies, case series, and systematic review articles published until October 2014 in the English or Spanish language, and evaluated their quality and the internal validity of the evidence provided. Two reviewers extracted the data independently, then both researchers simultaneously revised the data later, to arrive at a consensus. The search retrieved 1,158 papers, of which 965 were excluded for being duplicates, not relevant, or unrelated studies. A further 86 papers were excluded for being guidelines, protocols, or case reports, along with another 64 that did not contain relevant information, leaving 43 studies for inclusion. Many of these studies showed the utility of proteomic techniques for prediction, pathophysiology, diagnosis, management, monitoring, and prognosis of pre-eclampsia, perinatal infection, premature rupture of membranes, preterm birth, intrauterine growth restriction, and ectopic pregnancy. Proteomic techniques have enormous clinical significance and constitute an invaluable weapon in the management of obstetric disorders that increase maternal and perinatal morbidity and mortality.
Keywords: proteomic techniques, obstetrics, diagnosis, prediction
Introduction
The history of proteomics dates back to the 19th century and the studies of Gregor Johann Mendel (1822–1884), an Austrian monk and researcher whose observations and experiments using peas (1866) laid the foundations for our present-day laws of inheritance and genetics.1 As time passed, investigations into genetics and inheritance continued, and by 1930 researchers were able to grasp the true reach of Mendel’s results, heralding the start of the genomic era.1,2 Since that time, much genetic research has been undertaken in both plants and animals. In 2001, Collins and Venter announced to the world the culmination of the first erasers of human’s genetic map, which were finished two years later (2003), completing our genetic map.3,4
Genomics has contributed to advances in our understanding of human disease, but many aspects of pathology are still not understood, leading to the study of proteins as final products of gene expression, starting the proteomic era.5
Application of proteomics technology has had a great impact in terms of clarifying the etiology and pathogenesis of many diseases, and has contributed to the medical sciences via the development of methods for diagnosis, prognosis, and evaluation of treatment. Obstetrics is one of the branches of medicine that has benefited from these techniques. The aim of this review is to evaluate the utility of proteomic techniques in this specialty.
Methods
We searched the electronic databases of MEDLINE (US National Library of Medicine), EBSCOhost research databases, BVS Bireme, and SciELO, assisted by a clinical librarian, to identify the principal obstetric disorders that significantly increase maternal and perinatal morbidity and mortality in our region, including: “proteomics and pre-eclampsia”, “proteomics and perinatal infections”, “proteomics and premature rupture of membranes”, “proteomics and preterm birth”, “proteomics and intrauterine growth restriction”, “proteomics and ectopic pregnancy”, and “proteomics and pregnancy”. Other obstetric disorders were excluded.
Cohort studies, case-control studies, case series, and systematic reviews were considered for inclusion in our review. Included studies needed to have evaluated the utility of proteomics in the prediction, pathogenesis, diagnosis, treatment, or prognosis of pre-eclampsia, perinatal infection, premature rupture of membranes, preterm birth, intrauterine growth restriction, or ectopic pregnancy, and have been published up to October 2014 in the English or Spanish language. Guidelines, protocols, and case reports were excluded, and the internal validity of the evidence provided was checked. The two reviewers extracted the data independently, then both researchers simultaneously revised the data later, to arrive at a consensus.
Results and discussion
The MEDLINE search retrieved 891 records in English, the EBSCOhost search revealed a further 233 records in English and Spanish, the BVS Bireme search retrieved 32 studies in Spanish, and SciELO yielded two studies, also in Spanish.
In total, 965 papers were excluded for being duplicates, not relevant, or not related studies. We excluded a further 86 studies for being guidelines (n=11), study protocols (n=14), or case reports (n=61). We excluded another 64 papers that did not contain specific information on prediction, diagnosis, treatment, or prognosis, leaving 43 studies for inclusion in this review (Figure 1), 17 of which were cohort studies, 13 were reviews, ten were case-control studies, and three were case series.
  | Figure 1 Flowchart of studies. |
Definitions
In 1996, 7 years before publication of the human genetic map, research groups were starting to publish studies on proteins that were products of expression of the human genome, giving rise to the proteomic era. Wilkins et al defined proteomics as a union of techniques to explore simultaneously the presence and abundance of thousands of proteins in biological samples, including serum, blood, secretions, saliva, urine, cervical mucus, joint fluid, tissue fragments, and amniotic fluid.6
The very Wilkins et al exposed in other study, that proteomic has to do with a systematic analysis and in great scale of all proteins expressed by the genome of a cell, tissue, or organism in a specific moment and time conditions and environment that allow understanding of the sample composition, as well as the protein’s interaction in that sample.7
Peña-Castro et al described three techniques that could be used to study proteomics, ie, separation of proteins (by two-dimensional electrophoresis or multidimensional chromatography), identification of proteins (by mass spectrometry), and analysis and interpretation of the data obtained (by bioinformatics methods), and allow development of elements and the necessary algorithms to store and analyze large quantities of data concerning proteomes.8
In an earlier paper, Graves and Haystead observed that the proteome varies in space and time, unlike the genome, which is static in all cells within an organism,9 ie, two cells can have exactly the same sequence of DNA in the same organism, but the set of proteins expressed can be completely different depending not only on the cell type, but also on the biochemical environment surrounding the cell.
Medical applications
Proteins, being products of gene expression, have great potential as molecular biomarkers of disease, and are implicated in the majority of the physiological and pathological processes that take place in cells. They allow the discovery of useful markers for diagnosis, treatment, and monitoring of various clinical entities, and can even predict the optimal therapeutic strategy to use.10
According to Klein et al proteomics has had a great impact on the development of modern obstetrics, which contains multiple sources for the study of proteins, including maternal blood, maternal urine, placental tissue, amniotic fluid, cervical mucus, vaginal secretions, and saliva.11 The same researchers also pointed out that proteomics can be useful for prediction, diagnosis, management, monitoring, and prognosis of a number of obstetric conditions that are associated with an increased risk of maternal and/or perinatal morbidity and mortality (Figure 2), such as pre-eclampsia, perinatal infection, premature rupture of membranes, preterm birth, intrauterine growth restriction, and ectopic pregnancy.11
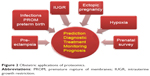  | Figure 2 Obstetric applications of proteomics. |
In 2002, Page et al reviewed a number of placental peptide markers associated with several obstetric conditions, including preterm delivery, intrauterine growth restriction, pre-eclampsia, and gestational diabetes, and underscored the value of proteomics in the diagnosis when estimating risk.12 In 2003, Kalionis and Moses reported similar applications for proteomics in research on pregnancy.13 The most important biomarkers identified are summarized in Table 1, as well as their specific utility in obstetrics.
  | Table 1 Summary of the most important biomarkers and their specific use in obstetric disorders |
Proteomics and pre-eclampsia
Pre-eclampsia is a disease of pregnant women that significantly increases maternal morbidity and mortality; its pathophysiology is unknown, but analysis of the proteome has been reported to be useful in this regard. Sawicki et al demonstrated that high levels of neurokinin B in cytotrophoblast cells are associated with the pathogenesis of pre-eclampsia, and that this protein suppresses the activity of other proteins like thioredoxin, cyclophilin A, cytokeratin 1, and peroxiredoxin 5, which play an important role in regulating cell antioxidant defenses and decrease levels of cytokeratin 1 and annexin 11, two proteins known to inhibit intravascular coagulation.14
A statistical analysis performed by Gharesi-Fard et al showed that chloride intracellular channel 3, apolipoprotein A-I, transthyretin, and protein disulfide isomerase are upregulated in the placental tissue of women with pre-eclampsia, with decreased expression of other proteins, including peroxiredoxin 2, peroxiredoxin 3, Cu/Zn-superoxide dismutase, actin gamma 1 propeptide, and chain A of enoyl-coenzyme A hydratase. These authors considered that all those protein dysfunctions can lead to disorders in antioxidant activities, playing a role in the pathogenesis of pre-eclampsia.15
Using the immunoassay technique, Watanabe et al showed that serum clusterin levels were significantly higher in pre-eclamptic women than in controls, but the presence of this protein alone was not sufficient to predict the disease in early pregnancy.16 Following on from this, Blumenstein et al monitored the activity of vitronectin and kininogen, starting several weeks before the onset of pre-eclampsia (gestational age 20 weeks) until the time that it was expressed, and observed a change in the normal proteolytic process for these two proteins in women with pre-eclampsia compared with controls, concluding that dysfunction in vitronectin and kininogen is useful for prediction of pre-eclampsia.17
A pilot study by Myers et al identified five proteins that were significantly upregulated in plasma samples taken at 26 weeks from pregnant women who later developed pre-eclampsia.18 Further, Carty et al identified 50 urinary biomarkers obtained at week 28 that were associated with future pre-eclampsia, reporting that fibrinogen alpha chain, collagen alpha chain, and uromodulin fragments were the most important early predictors of pre-eclampsia.19
Specific fragments of Serpina-1 and albumin in urine samples have also been reported to be unique proteomic markers for prediction of pre-eclampsia, and can be used to make the differential diagnosis out of the hypertensive disorders occurring in pregnant women. Of these, the 21 amino acid C-terminal fragment of Serpina-1 is significantly associated with severe forms of pre-eclampsia.20 Likewise, Sun et al identified trophoblastic proteins associated with pre-eclampsia, reporting that disulfide isomerase ER-60, peroxiredoxin 2, and delta 3,5-delta 2,4-dienoyl-CoA isomerase were significantly repressed in patients with pre-eclampsia, while protein disulfide isomerase precursor, endoplasmic reticulum resident protein, dihydrolipoyl dehydrogenase, and TIM21-like protein were upregulated.21
In a recent study reported by Ma et al significant differences were found in 70 proteins within the trophoblastic cells of women with pre-eclampsia when compared with healthy control women. Thirty-one of these proteins were downregulated and 39 were upregulated, and these findings could help in the early diagnosis of pre-eclampsia.22
Proteomics, perinatal infections, and preterm birth
Paiva et al have identified several serum biomarkers found in the different stages of sepsis and involved with the immune system, in particular protease inhibitor peptides. Their findings also suggest that the complement and coagulation pathways related to lipid metabolism participate in sepsis.23
Preterm birth is commonly caused by intra-amniotic infection. Gravett et al performed a study in rhesus monkeys to identify protein markers before and after experimental intra-amniotic infection. They then investigated peptide profiles in women who delivered prematurely with or without intra-amniotic infection and in women who delivered at term. This study identified calgranulin B and a fragment of insulin-like growth factor binding protein 1 (IGFBP-1) as useful biomarkers for early detection of intra-amniotic infection.24 Another study by the same group of investigators analyzed the proteome of cervical–vaginal fluid in primates with experimentally induced intra-amniotic Ureaplasma parvum infection, and identified potential biomarkers such as calgranulins A and B, IGFBP-1, azurocidin, lipocalin, annexin II, and an unregulated protein (haptoglobin) to be associated with inflammatory and defense responses.25
Rüetschi et al identified human neutrophil proteins 1, 2 and 3, and calgranulins A and B in amniotic and cervical fluid samples from women with preterm labor.26 Pereira et al also observed the presence of calgranulins, annexins, and epidermal fatty acid binding protein in cervical–vaginal fluid from patients with preterm labor and spontaneous preterm birth, but also found other proteins, like alpha-1-antitrypsin, haptoglobin, and serotransferrin, in serum samples from such patients.27 In another study, the same research team found that proteins involved in preterm labor can be classified into four functional groups, ie, complement or coagulation cascade, immune response, fetal and placental development, and extracellular matrix proteins.28
According to Buhimschi et al there are four biomarkers in amniotic fluid that can predict perinatal infection, ie, human neutrophil defensins 1 and 2 and calgranulins A and C.29,30 Buhimschi et al have also reported that proteomic biomarkers of intra-amniotic inflammation emerge sequentially in the process of inflammation and infection, starting with human neutrophil defensin 2, followed by human neutrophil defensin 1 and calgranulin C, and ending with calgranulin A. These authors observed that the presence of these four components confirms fetal and/or neonatal infection, and that these biomarkers are more reliable than other methods for diagnosis of infection and preterm birth.31 Buhimschi et al also showed that proteomic study of amniotic fluid samples can identify patients at high risk for preterm delivery, even without intra-amniotic infection or bleeding, and determined that distinctive proteins involved in signal transport, transduction, and metabolism are expressed in high risk patients.32
Liong et al recently identified proteins in cervical–vaginal fluid that could be useful for predicting preterm labor in asymptomatic women. They found that thioredoxin and interleukin-1 receptor antagonist were significantly reduced for up to 90 days before onset of preterm labor when compared with women who delivered at term. Both markers showed a high positive and negative predictive value.33 Further, in a recent systematic review, Nápoles Méndez reminded us of other classic proteomic markers described in the literature than help to predict preterm birth, such as alpha-fetoprotein, C-reactive protein, interleukin-1, interleukin-6, and interleukin-8, tumor necrosis factor, matrix metalloproteinase-8, and fetal fibronectin.34
Proteomics and intrauterine growth restriction
A study performed by Karamessinis et al reported changes at the post-translational modification level in alpha-2-HS glycoprotein (fetuin-A) protein sampled from umbilical cord plasma in intrauterine growth restriction cases. This glycoprotein is closely linked to fetal growth, cell replication, and osteogenesis.35
Another protein involved in fetal growth and development is IGFBP-1, and recent data show that some phosphorylation sites can be assessed in human amniotic fluid samples in order to clarify the potential mechanism of fetal growth.36 Analysis of plasma samples from pregnant women also allows detection of several proteins, including Serpina-3 and antichymotrypsin, which are depleted in cases of intrauterine growth restriction complicated by pre-eclampsia.37
A study by Cecconi et al identified 25 proteins expressed in patients with intrauterine growth restriction (14 in umbilical cord serum samples and 11 in amniotic fluid samples). Another finding in this study was that some of these proteins were involved in the immune and inflammatory response and others were involved in coagulation, transport, and blood pressure regulation, all of which play a role in the pathogenesis of intrauterine growth restriction.38
A proteome study by Wölter et al found that apolipoprotein C-III is more abundant in cord blood serum samples from patients with intrauterine growth restriction independent of gestational age.39 Further, according to Ferrara and Viel, there are other useful proteomic biomarkers for diagnosis of intrauterine growth restriction, including leptin, transforming growth factor, fibroblast growth factor, ectodermal growth factor, and platelet-derived growth factor.40
Proteomics and ectopic pregnancy
Gerton et al identified several serum proteins in the 7,500–18,000 Da mass range that can distinguish an ectopic pregnancy from a normal intrauterine pregnancy. Based on sensitivity and specificity values, these authors developed two algorithms that classify patients into three main groups, ie, patients who need immediate intervention for a probable ectopic pregnancy, patients with a normal pregnancy, and patients who need monitoring to make the diagnosis.41
A case-control study performed by Rausch et al found a significant decrease in serum matrix metalloproteinase-12 levels in women with ectopic pregnancy, with a sensitivity of 97%. Nevertheless, the authors considered that further studies are needed to confirm the diagnostic value of this biomarker.42
Evidence to be considered
In view of the evidence presented here, we are concerned that according to the report by Anderson,43 only 205 proteins from approximately 211 genes are available for clinical tests, providing quantitative measurements for approximately 1% of the human protein gene products. New protein analytes has remained essentially flat over the past 15 years, averaging 1.5 new proteins per year.43
Conclusion
Proteomic techniques are invaluable with regard to prediction, pathophysiology, early diagnosis, timely treatment, and prognosis of many of the obstetric disorders than increase maternal and perinatal morbidity and mortality. We therefore ask ourselves if the proteomic and/or other “-omic” techniques can answer our questions? Are the molecular biochemistry, the genetics, and the immunology, the principal weapons to understand the diseases behavior in the future? We believe so.
Disclosure
The authors report no conflicts of interest in this work.
References
Gomis A. [History for Scientists]. Madrid, Spain: Ediciones Nivola; 2000. Spanish. | ||
Sturtevant AH. A History of Genetics. New York, NY, USA: Harper & Row; 1965. | ||
Lander ES, Linton LM, Birren B, et al; International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. | ||
Flanagan K, Stevens R, Pocock M, Lee P, Wipat A. Ontology for genome comparison and genomic rearrangements. Comp Funct Genomics. 2004;5:537–544. | ||
Manolio TA, Collins FS. The HapMap and genome-wide association studies in diagnosis and therapy. Annu Rev Med. 2009;60:443–456. | ||
Wilkins MR, Sanchez JC, Gooley AA, et al. Progress with proteome projects: why all proteins expressed by a genome should be identified and how to do it. Biotechnol Genet Eng Rev. 1996;13:19–50. | ||
Wilkins MR, Pasquali C, Appel RD, et al. From proteins to proteomes: large scale protein identification by two-dimensional electrophoresis and amino acid analysis. Biotechnology (N Y). 1996;14:61–65. | ||
Peña-Castro JM, Gregorio-Ramírez O, Barrera-Figueroa BE. [Experimental Methods Used to Study Macromolecules: History, Rationale and Prospects]. Educ Quím. 2013;24:237–246. Spanish. | ||
Graves PR, Haystead TA. Molecular biologist’s guide to proteomics. Microbiol Mol Biol Rev. 2002;66:39–63. | ||
Jara A, Kopchick JJ. [Proteomics: An Integrated Approach]. An Pediatr (Barc). 2013;78:137–139. Spanish. | ||
Klein J, Buffin-Meyer B, Mullen W, et al. Clinical proteomics in obstetrics and neonatology. Expert Rev Proteomics. 2014;11:75–89. | ||
Page NM, Kemp CF, Butlin DJ, Lowry PJ. Placental peptides as markers of gestational disease. Reproduction. 2002;123:487–495. | ||
Kalionis B, Moses E. Advanced molecular techniques in pregnancy research: proteomics and genomics – a workshop report. Placenta. 2003;24 Suppl A:S119–S122. | ||
Sawicki G, Dakour J, Morrish DW. Functional proteomics of neurokinin B in the placenta indicates a novel role in regulating cytotrophoblast antioxidant defences. Proteomics. 2003;3:2044–2051. | ||
Gharesi-Fard B, Zolghadri J, Kamali-Sarvestani E. Proteome differences of placenta between preeclampsia and normal pregnancy. Placenta. 2010;31:121–125. | ||
Watanabe H, Hamada H, Yamada N, et al. Proteome analysis reveals elevated serum levels of clusterin in patients with preeclampsia. Proteomics. 2004;4:537–543. | ||
Blumenstein M, Prakash R, Cooper GJ, North RA; SCOPE Consortium. Aberrant processing of plasma vitronectin and high molecular weight kininogen precedes the onset of preeclampsia. Reprod Sci. 2009;16:1144–1152. | ||
Myers J, Macleod M, Reed B, Harris N, Mires G, Baker P. Use of proteomic patterns as a novel screening tool in pre-eclampsia. J Obstet Gynaecol. 2004;24:873–874. | ||
Carty DM, Siwy J, Brennand JE, et al. Urinary proteomics for prediction of preeclampsia. Hypertension. 2011;57:561–569. | ||
Buhimschi IA, Zhao G, Funai EF, et al. Proteomic profiling of urine identifies specific fragments of Serpina-1 and albumin as biomarkers of preeclampsia. Am J Obstet Gynecol. 2008;199:551.e1–e16. | ||
Sun LZ, Yang NN, De W, Xiao YS. Proteomic analysis of proteins differentially expressed in preeclamptic trophoblasts. Gynecol Obstet Invest. 2007;64:17–23. | ||
Ma K, Jin H, Hu R, et al. A proteomic analysis of placental trophoblastic cells in preeclampsia-eclampsia. Cell Biochem Biophys. 2014;69:247–258. | ||
Paiva RA, David CM, Domont GB. Proteomic and sepsis: a pilot study. Rev Bras Ter Intensiva. 2010;22:403–412. | ||
Gravett MG, Novy MJ, Rosenfeld RG, et al. Diagnosis of intra-amniotic infection by proteomic profiling and identification of novel biomarkers. JAMA. 2004;292:462–469. | ||
Gravett MG, Thomas A, Schneider KA, et al. Proteomic analysis of cervical-vaginal fluid: identification of novel biomarkers for detection of intra-amniotic infection. J Proteome Res. 2007;6:89–96. | ||
Rüetschi U, Rosén A, Karlsson G, et al. Proteomic analysis using protein chips to detect biomarkers in cervical and amniotic fluid in women with intra-amniotic inflammation. J Proteome Res. 2005;4:2236–2242. | ||
Pereira L, Reddy AP, Jacob T, et al. Identification of novel protein biomarkers of preterm birth in human cervical-vaginal fluid. J Proteome Res. 2007;6:1269–1276. | ||
Pereira L, Reddy AP, Alexander AL, et al. Insights into the multifactorial nature of preterm birth: proteomic profiling of the maternal serum glycoproteome and maternal serum peptidome among women in preterm labor. Am J Obstet Gynecol. 2010;202:555.e1–e10. | ||
Buhimschi IA, Christner R, Buhimschi CS. Proteomic biomarker analysis of amniotic fluid for identification of intra-amniotic inflammation. BJOG. 2005;112(2):173–181. | ||
Buhimschi CS, Buhimschi IA, Abdel-Razeq S, et al. Proteomic biomarkers of intra-amniotic inflammation: relationship with funisitis and early-onset sepsis in the premature neonate. Pediatr Res. 2007;61:318–324. | ||
Buhimschi CS, Dulay AT, Abdel-Razeq S, et al. Fetal inflammatory response in women with proteomic biomarkers characteristic of intra-amniotic inflammation and preterm birth. BJOG. 2009;116:257–267. | ||
Buhimschi IA, Zhao G, Rosenberg VA, Abdel-Razeq S, Thung S, Buhimschi CS. Multidimensional proteomics analysis of amniotic fluid to provide insight into the mechanisms of idiopathic preterm birth. PLoS One. 2008;3:e2049. | ||
Liong S, Di Quinzio MK, Fleming G, Permezel M, Rice GE, Georgiou HM. Prediction of spontaneous preterm labour in at-risk pregnant women. Reproduction. 2013;146:335–345. | ||
Nápoles Méndez D. [Some proteomic markers in perinatal medicine]. Rev Cubana Obstet Ginecol. 2013;39:209–222. Spanish. | ||
Karamessinis PM, Malamitsi-Puchner A, Boutsikou T, et al. Marked defects in the expression and glycosylation of alpha2-HS glycoprotein/fetuin-A in plasma from neonates with intrauterine growth restriction: proteomics screening and potential clinical implications. Mol Cell Proteomics. 2008;7:591–599. | ||
Abu Shehab M, Inoue S, Han VK, Gupta MB. Site specific phosphorylation of insulin-like growth factor binding protein-1 (IGFBP-1) for evaluating clinical relevancy in fetal growth restriction. J Proteome Res. 2009;8:5325–5335. | ||
Auer J, Camoin L, Guillonneau F, et al. Serum profile in preeclampsia and intra-uterine growth restriction revealed by iTRAQ technology. J Proteomics. 2010;73:1004–1017. | ||
Cecconi D, Lonardoni F, Favretto D, et al. Changes in amniotic fluid and umbilical cord serum proteomic profiles of fetuses with intrauterine growth retardation. Electrophoresis. 2011;32:3630–3637. | ||
Wölter M, Röwer C, Koy C, et al. A proteome signature for intrauterine growth restriction derived from multifactorial analysis of mass spectrometry-based cord blood serum profiling. Electrophoresis. 2012;33:1881–1893. | ||
Ferrara SD, Viel G. Functional ‘-omics’ in intrauterine growth restriction: novel insights into child development. Expert Rev Proteomics. 2012;9:355–357. | ||
Gerton GL, Fan XJ, Chittams J, et al. A serum proteomics approach to the diagnosis of ectopic pregnancy. Ann N Y Acad Sci. 2004;1022:306–316. | ||
Rausch ME, Beer L, Sammel MD, et al. A disintegrin and metalloprotease protein-12 as a novel marker for the diagnosis of ectopic pregnancy. Fertil Steril. 2011;95:1373–1378. | ||
Anderson NL. The clinical plasma proteome: a survey of clinical assays for proteins in plasma and serum. Clín Chem. 2010;56:177–185. |
 © 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
