Back to Journals » Pharmacogenomics and Personalized Medicine » Volume 10
Urine metabolic ratio of omeprazole in relation to CYP2C19 polymorphisms in Russian peptic ulcer patients
Authors Denisenko NP , Sychev DA, Sizova ZM, Smirnov VV, Ryzhikova KA , Sozaeva ZA, Grishina EA
Received 16 May 2017
Accepted for publication 31 August 2017
Published 27 September 2017 Volume 2017:10 Pages 253—259
DOI https://doi.org/10.2147/PGPM.S141935
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Martin H Bluth
Natalia P Denisenko,1–3 Dmitriy A Sychev,2 Zhanna M Sizova,3 Valeriy V Smirnov,4,5 Kristina A Ryzhikova,1 Zhannet A Sozaeva,1 Elena A Grishina1
1Research Center, Russian Medical Academy of Continuous Professional Education, Ministry of Healthcare, Moscow, Russia; 2Department of Clinical Pharmacology and Therapy, Russian Medical Academy of Continuous Professional Education, Ministry of Healthcare, Moscow, Russia; 3Department of Social Expertise, Urgent and Outpatient Therapy, First Moscow State Medical University (Sechenov University), Ministry of Healthcare, Moscow, Russia; 4Department of Pharmaceutical and Toxicological Chemistry, First Moscow State Medical University (Sechenov University), Ministry of Healthcare, Moscow, Russia; 5Laboratory of Clinical Pharmacology, National Research Centre, Institute of Immunology, Federal Medical Biological Agency, Moscow, Russia
Background: CYP2C19 is known to be the main enzyme of biotransformation of proton pump inhibitors (PPIs), whereas the CYP2C19 gene is highly polymorphic. Genotyping and phenotyping together represent more reliable data about patient’s CYP2C19 activity.
Purpose: The aim of the study was to investigate the applicability of urine metabolic ratio of omeprazole for CYP2C19 phenotyping in Russian peptic ulcer patients with different CYP2C19 genotypes.
Patients and methods: A total of 59 patients (19 men and 40 women) aged 18–91 years (mean age 53.5±15.1 years) from four Moscow clinics who were diagnosed with an endoscopically and histologically proven peptic ulcer or had a history of endoscopically and histologically proven ulcers in the past were recruited. Peripheral venous blood (6 mL) was collected for DNA extraction, and real-time polymerase chain reaction was performed for the analysis of CYP2C19*2G681A (rs4244285), CYP2C19*3G636A (rs4986893) and CYP2C19*17C-806T (rs12248560) polymorphisms. Urine samples of patients were collected in the morning between 6 am and 9 am, before food or drug intake, after at least 3 days of twice daily (b.i.d.) omeprazole intake. Omeprazole and 5-hydroxyomeprazole concentrations in the urine were measured using high-performance liquid chromatography with mass spectrometry.
Results: Of the 59 patients, there were 27 (45.8%) extensive metabolizers (EMs; CYP2C19*1/*1), 16 (27.1%) ultrarapid metabolizers (UMs; CYP2C19*1/*17, CYP2C19*17/*17), 14 (23.7%) intermediate metabolizers (IMs; CYP2C19*1/*2, CYP2C19*2/*17, CYP2C19*3/*17) and two (3.4%) poor metabolizers (PMs; CYP2C19*2/*2). Median metabolic ratio (25%–75% percentiles) were 1.03 (0.69–1.36) for EMs, 1.95 (1.33–2.68) for UMs, 1.40 (0.78–2.13) for IMs+PMs and 1.26 (0.82–1.99) for the whole sample. A statistically significant difference in metabolic ratio (Mann–Whitney U test) was found between UMs and EMs (p=0.001) and in the multiple comparison Kruskal–Wallis test (p=0.005).
Conclusion: We found a connection between particular CYP2C19 genotypes and urine metabolic ratio of omeprazole in Russian peptic ulcer patients. This method needs to be improved as in our modification it worked mainly for UMs and did not differentiate all patients according to omeprazole biotransformation activity.
Keywords: pharmacogenetics, phenotyping, metabolomics, proton pump inhibitor
Introduction
Cytochrome P450 (CYPs) is a group of Phase I metabolizing enzymes that are widely involved in biotransformation of xenobiotics.1 Cytochrome P 450 2C19 isoenzyme (CYP2C19) is known to be the main enzyme of biotransformation of proton pump inhibitors (PPIs), whereas CYP2C19 gene is highly polymorphic. CYP2C19*2 and CYP2C19*3 polymorphisms found in 1994 and CYP2C19*17 polymorphism found in 2006 have proved to influence the efficacy of treatment of peptic ulcer patients.2–4 CYP2C19*2 and CYP2C19*3 carriers were proved to have slow biotransformation of PPIs, whereas CYP2C19*17 carriers are suggested to have rapid metabolism of PPIs that may result in insufficient drug response.5
Dutch Pharmacogenetics Working Group of the Royal Dutch Pharmacists Association developed guidelines, which translate the results of pharmacogenetic tests to specific therapeutic recommendations.6 These guidelines are based on systematic review of literature and represent dose recommendations for different polymorphism carriers. On the basis of the guidelines, CYP2C19 polymorphisms’ carriers are divided into extensive metabolizers (EMs), intermediate metabolizers (IMs), poor metabolizers (PMs) and ultrarapid metabolizers (UMs). These are predicted phenotypes of CYP2C19 based on CYP2C19 genotype.
CYP2C19 phenotyping complements genotyping. Genotyping and phenotyping together represent more reliable data about patient’s CYP2C19 activity. S-Mephenytoin was a probe drug for CYP2C19 activity in urine in the past but because of its potential adverse effects, omeprazole was offered. Studies showed that omeprazole hydroxylation index in plasma correlates with S-mephenytoin metabolism in urine.7,8
Correlation between CYP2C19 genotype and omeprazole metabolic ratio (5-hydroxyomeprazole/omeprazole) or hydroxylation index (omeprazole/5-hydroxyomeprazole) was shown by Ohnishi et al.9
We conducted this study to investigate the applicability of urine metabolic ratio of omeprazole for CYP2C19 phenotyping in Russian peptic ulcer patients with different CYP2C19 genotypes.
Patients and methods
Study design and patients
Patients from four Moscow clinics who were diagnosed with gastric or duodenal peptic ulcer based on endoscopy and histology (exacerbation) or had medical records with documented endoscopically and histologically proven ulcers in the past (remission) and who underwent gastroscopy and were taking omeprazole (either for ulcer treatment or concomitant erosive gastritis, duodenitis, gastroesophageal reflux disease treatment or gastric protection) were recruited. The inclusion criteria were: had been diagnosed with peptic ulcer (exacerbation or remission) and omeprazole 20 mg twice daily (b.i.d.) prescribed for at least 7 days intake. The exclusion criteria were people younger than 18 years, omeprazole hypersensitivity, other than omeprazole PPI prescribed or change in PPI during treatment, severe comorbidities (cardiovascular, cerebral, renal, hepatic dysfunction), pregnancy and lactation, and refusal of participation in any part of the study. The study was approved by local ethics committee of First Moscow State Medical University (Sechenov University). All patients signed written informed consent to participate in the study.
A total of 59 peptic ulcer patients (19 men and 40 women) of Russian nationality from Moscow region aged 18–91 years (mean age 53.5±15.1 years) were enrolled.
All patients had gastrointestinal complains: dyspepsia, abdominal pain and heartburn. According to the results of gastroscopy, 18 (30.5%) patients had acute peptic ulcer and 41 (69.5%) patients who had endoscopically and histologically proven history of peptic ulcer were diagnosed with erosive gastritis, erosive duodenitis, gastroesophageal reflux disease, Helicobacter pylori infection (rapid urease test and histological or cytological examinations, 10 patients) separately or in combination and ulcer remission when enrolled to the study. Omeprazole was prescribed to all the patients by attending physicians; dose regimen was 20 mg b.i.d. Omeprazole brand names differed: 27 (45.8%) patients were taking omeprazole (OAO Synthesis, Kurgan, Russia), 13 (22.0%) omeprazole (OOO Promed, Rostov region, Aksai, Russia), 12 (20.3%) Omez (Dr. Reddy’s Laboratories Ltd., Bachupally Village, Telangana State, India) and 7 (11.9%) omeprazole-Teva (Teva Pharmaceutical Industries Ltd., Petach Tikva, Israel). Drug inhibitors or inducers of CYP2C19 enzyme were not prescribed to patients.
Genotyping
Peripheral venous blood (6 mL) of patients was collected in K2-ethylenediaminetetraacetic acid (EDTA) tubes (DNK; Sunphoria Co., Ltd., Taipei City, Taiwan) for DNA extraction from leukocytes. Blood samples were stored deeply frozen to -70°C until analysis. DNA was extracted using commercially available kit with silica sorbent (Syntol, Moscow, Russia). The kit consisted of lysis solution (cell lysis reagent), sorbent for DNA sorption, washing solutions and solution for DNA elution. There were four steps for DNA extraction: cell lysis in the sample, DNA sorption, washing until sorbent was resuspended and DNA elution and supernatant collection.
CYP2C19*2G681A (rs4244285), CYP2C19*3G636A (rs4986893) and CYP2C19*17C-806T (rs12248560) polymorphisms were analyzed using real-time polymerase chain reaction (PCR). PCR was performed using commercially available kit (produced by DNA-Technology, Moscow, Russia) in DNA amplifier DTlite (DNA-Technology). The kit consisted of three amplification mixtures (CYP2C19: 681 G>A *2, CYP2C19: 636 G>A *3, CYP2C19: -806C>T *17), PCR buffer, Taq polymerase and mineral oil. The program consisted of initial denaturation step at 95°C for 3 minutes followed by 40 cycles of 15 seconds’ denaturation at 95°C and then annealing at 63°C for 40 seconds. Genotyping was repeated for 20% of patients’ samples as a control for correct sample handling. Repeated genotyping revealed identical results. Patients were referred to as CYP2C19 EMs, PMs, IMs or UMs according to the Dutch Pharmacogenetics Working Group Guideline of the Royal Dutch Pharmacists Association.6
Phenotyping
Urine samples of patients were collected in the morning between 6 and 9 am before food or drug intake after at least three days of b.i.d. omeprazole intake on the fourth day or later. This time was chosen empirically for the reason that five elimination half-life periods of omeprazole were completed and steady-state concentration was thought to be achieved. We evaluated metabolic ratio of 5-hydroxyomeprazole/omeprazole in urine, which is suggested to reflect the activity of CYP2C19 to metabolize omeprazole.
Urine samples (10.5 mL) were collected in plastic tubes (RusTech, Moscow, Russia) without reagents and were stored deeply frozen to -70°C until analysis. Omeprazole and 5-hydroxyomeprazole concentrations in urine were measured using Agilent 1290 Infinity (Agilent Technologies, Santa Clara, CA, USA) high-performance liquid chromatography with mass spectrometry (HPLC–MS). The HPLC–MS method was developed and validated. The isolation of drug and its metabolite was performed using Agilent Eclipse XDB-C18 Column (50 mm×2.1 mm; 1.8 μm). The column temperature was maintained at 50°C. The mobile phase A was 5 mM ammonium formate in 0.01% water formic acid solution, and the mobile phase B was 0.01% acetonitrile formic acid solution. The flow rate of both mobile phases was 0.4 mL/min. Gradient elution is described in Table 1.
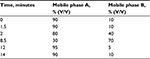  | Table 1 Gradient elution for HPLC–MS Abbreviation: HPLC–MS, high-performance liquid chromatography with mass spectrometry. |
Volumes of 10 μL were injected. Retention times were 3.5, 4.3, and 7.2 minutes for omeprazole, 5-hydroxyomeprazole, and the internal standard carbamazepine, respectively. The total HPLC run time was 14 minutes. The mass spectrometer was operated using the following conditions: electrospray ionization (ESI), capillary voltage 3500 V, ion tube temperature 350°C, ionization camera temperature 400°C, drying gas (nitrogen) flow rate 10 L/min and sheath gas flow 11 L/min.
The HPLC–MS parameters are listed in Table 2.
  | Table 2 Analytical parameters for HPLC–MS Abbreviation: HPLC–MS, high-performance liquid chromatography with mass spectrometry. |
Urine metabolic ratios were calculated as the ratio of the amount of metabolite (5-hydroxyomeprazole) to that of substrate (omeprazole) in urine. The higher the urine metabolic ratio, the higher the activity of CYP2C19.
Statistical analysis
The statistical analysis was performed using SPSS Statistics 22 (IBM Corporation, Armonk, NY, USA). A p-value <0.05 was considered as statistically significant. The normality of metabolic ratio distribution was analyzed by the Kolmogorov–Smirnov test. Nonparametric statistics was used to compare metabolic ratio in different CYP2C19 genotypes. The Kruskal–Wallis test was used for comparisons between three CYP2C19 genotype groups. The observed and expected genotype frequencies were calculated by using the Hardy–Weinberg equilibrium. Fisher’s exact test was used to evaluate deviation of genotype frequencies in the studied population from the Hardy–Weinberg equilibrium.
Results
Of 59 patients, there were 27 (45.8%) EMs (CYP2C19*1/*1), 16 (27.1%) UMs (CYP2C19*1/*17, CYP2C19*17/*17), 14 (23.7%) IMs (CYP2C19*1/*2, CYP2C19*2/*17, CYP2C19*3/*17) and 2 (3.4%) PMs (CYP2C19*2/*2). Genotype frequencies are reported in Table 3.
  | Table 3 Frequency of CYP2C19 polymorphisms in peptic ulcer patients Abbreviations: EMs, extensive metabolizers; IMs, intermediate metabolizers; PMs, poor metabolizers; UMs, ultrarapid metabolizers. |
All genotypes were in accordance to the Hardy–Weinberg equilibrium (p>0.05; Table 4). The allele frequencies were as follows: CYP2C19*2 – 14.4%, CYP2C19*3 – 0.8% and CYP2C19*17 – 17.8%.
  | Table 4 Frequency of CYP2C19 genotype in peptic ulcer patients |
PMs and IMs were combined in one group for further analysis.
Because distribution of metabolic ratio was not normal (Kolmogorov–Smirnov test, p=0.038), median and 25%–75% percentiles for data description and nonparametric statistics for samples’ comparison were used (Table 5).
Metabolic ratio of omeprazole was the highest in CYP2C19*1/*17 patient (3.24) and the lowest in CYP2C19*1/*1 patient (0.26). Urine metabolic ratio distribution in patients with different CYP2C19 genotypes is shown in Figure 1.
A statistically significant difference in metabolic ratio (Mann–Whitney U test) was found between UMs and EMs (p=0.001) and in multiple comparison Kruskal–Wallis test (p=0.005; Table 5).
There were no relation between omeprazole brand name and urine metabolic ratio (Fisher’s exact test, p>0.05). There were no relation between the day of urine sample collection (fourth or later day) and urine metabolic ratio (Fisher’s exact test, p>0.05).
Discussion
The frequency of CYP2C19*2, CYP2C19*3 and CYP2C19*17 alleles of Russian peptic ulcer patients in our study is comparable with data described for Europeans.10
CYP2C19*3 occurs in <1% of Caucasians (0.8% in our study) and is more common in Asians.11,12 CYP2C19*2 is rather common, and its frequency is near 15% in the European population and 14.4% in our patients.13–15 Clinical significance of CYP2C19*17 was found in 2006; its frequency is ~20% in different populations and is still under research.4 We found 17.8% CYP2C19*17 allele frequencies in our study.
Plasma hydroxylation index of omeprazole (omeprazole/5-hydroxyomeprazole) after 3 hours of 20 mg omeprazole single-dose intake in Iranian study with 180 healthy volunteers was studied.16 Mean hydroxylation index was 0.35±0.06 for CYP2C19*17/*17 carriers, 0.75±0.28 for CYP2C19*1/*17 carriers, 0.85±0.30 for CYP2C19*1/*1 carriers, 2.02±0.84 for CYP2C19*2/*17 carriers, 2.27±1.04 for CYP2C19*1/*2 carriers and 13.59±3.13 for CYP2C19*2/*2 carriers. Thus, investigators found the correlation between CYP2C19 genotype and phenotype. Median hydroxylation index statistically significantly differed in CYP2C19*17/*17 compared with other CYP2C19 genotypes. CYP2C19*2/*2 median hydroxylation index had statistically significant difference compared with other CYP2C19 genotypes. Investigators concluded that phenotyping helped to define CYP2C19*17/*17 UMs and CYP2C19*2/*2 PMs. Omeprazole hydroxylation index is reverse to metabolic ratio studied in our work; if translated, metabolic ratios for EMs and UMs are comparable with our data.
There is lack of phenotyping methods described in previous literature. Many studies include a 6–12 hour-period for blood and urine samples collection for pharmacokinetics and were relatively high in costs and hard for application and therefore had limited introduction to practice. The number of participants in such studies was rather low because of complicated design.
In Iranian study with 30 healthy volunteers after 20 mg omeprazole single dose, blood samples were collected for 12 hours and omeprazole and 5-hydroxyomeprazole concentrations for pharmacokinetics were measured.17 Urine concentrations were not studied.
In some other studies, omeprazole concentration in urine could not be measured because of unsuitable buffer that caused omeprazole degradation in urine samples.18,19
Nevertheless, some studies succeed in omeprazole and 5-hydroxyomeprazole determination in urine. In Finnish study, blood and urine samples were taken for 6-hour period from four volunteers after oral cocktail (melatonin, nicotine, bupropion, repaglinide, losartan, omeprazole, dextromethorphan, chlorzoxazone, midazolam) intake.20 Drugs and their metabolites were measured for CYPs phenotyping using the liquid chromatography tandem–mass spectrometry (LC-MS/MS) method. Blood omeprazole and 5-hydroxyomeprazole samples were used for CYP2C19 phenotyping, whereas urine samples with all time points were acquired from only two volunteers. Volunteers after 10 mg omeprazole single dose had the following omeprazole and metabolite pharmacokinetics: area under the curve (AUC) 0–6 hours: 552.5 and 396.5 ng/mL⋅h, cmax=158.4±113.2 ng/mL and 109.2±62.9 ng/mL, tmax=4 and 4 hours, cumulative excretion in urine 5.7–119.0 µg and 38.3–4056.0 µg and maximum excretion rate in urine 26.3–51.1 µg/h and 649.0–1392.0 µg/h.
Turkish authors studied CYP2C19 metabolic activity using lansoprazole to 5-hydroxylansoprazole in plasma in healthy children with determined CYP2C19 polymorphisms. They found that mean plasma concentration of lansoprazole was 70% lower in CYP2C19*17/*17 UMs compared with CYP2C19*1/*1 EMs (p<0.05). Mean plasma concentration of lansoprazole was 6.9 times higher in CYP2C19*2/*2 PMs compared with that in CYP2C19*1/*1 EMs (p<0.01). In CYP2C19*17/*17 UMs, lansoprazole/5-hydroxylansoprazole ratio was significantly lower compared with that in CYP2C19*1/*1 EMs (2.8±2.1 vs 6.1±4.5). CYP2C19*2/*2 PMs had significantly higher lansoprazole metabolic ratio compared with CYP2C19*1/*1 EMs (63.5±12.2 vs 6.1±4.5). The authors concluded that CYP2C19*2 and CYP2C19*17 are important for prognosing clinical outcomes, and lansoprazole may be used as a probe drug for CYP2C19 phenotyping.21
One time point and optimal biomatrix for phenotyping were studied in the following work. The goal of investigators was to find optimal matrix (plasma or urine) and one time point for measuring drugs and metabolites from cocktail (caffeine, losartan, omeprazole, and dextromethorphan) to reflect the activity of CYPs.19 Blood and urine samples were taken for 12 hours period from 13 healthy volunteers for CYP2C19 phenotyping. As for plasma metabolic ratio (omeprazole AUC/5-hydroxyomeprazole AUC), optimal time point was 3–12 hours after 20 mg omeprazole single dose. For urine metabolic ratio, investigators failed to determine optimal time point as omeprazole concentration in urine was not identified for some volunteers.
Hydroxylation index (omeprazole AUC/5-hydroxyomeprazole AUC) based on 3 hours after omeprazole intravenous or oral administration was optimal for CYP2C19 activity estimation in a Japanese study.22
We suggest that phenotyping methods based on single blood or urine intake are more applicable for clinicians. Collection of blood samples for phenotyping is invasive and needs equipment for the following plasma centrifugation. These limitations are avoided for urine samples as a biomatrix for phenotyping, thus making the method more simple for clinical practice. It is also obvious that drug and metabolite concentrations in urine are less precise than those in plasma, mostly because of partial intestinal excretion. Metabolic ratio of metabolite concentration to drug concentration is suggested to reduce this imperfection. No optimal time point was found for collecting urine samples for CYP2C19 phenotyping in literature. We offered phenotyping method based on long-term omeprazole intake with urine sample collection in morning on the fourth or later day of b.i.d. omeprazole intake.
The main limitations of the study are small sample size, absence of plasma omeprazole and 5-hydroxyomeprazole concentrations and one CYP isoenzyme detection both in genotyping and phenotyping. Other CYPs such as CYP3A4, CYP3A5, CYP1A2 may also influence the results of the study. In conditions of reduced CYP2C19 activity, PPI metabolism may go through other CYPs. Induction of CYP1A2 in multiple omeprazole dosing was also shown in previous studies.23,24
Conclusion
We found a connection between particular CYP2C19 genotypes and urine metabolic ratio of omeprazole in Russian peptic ulcer patients with long-term omeprazole intake. The data need to be verified in further studies on considerable samples. Urine concentrations of omeprazole as a probe drug for CYP2C19 isoenzyme and its CYP isoform-specific metabolite 5-hydroxyomeprazole are promising for indicating CYP2C19 enzyme activity. This method needs to be improved as in our modification it works mainly for UMs and does not differentiate all patients according to omeprazole biotransformation activity. CYP2C19 phenotyping needs complete registration of concomitant drugs, herbals and biologically active additives, components of which may inhibit or induce CYP2C19 enzyme.
Acknowledgments
This work was supported by the Russian Scientific Foundation (project number 16-15-00227: “Fundamental research and exploratory research in priority areas of research”) for CYP2C19*2 and CYP2C19*3 genotyping reagents and consumables and phenotyping reagents and consumables and the University Grant of First Moscow State Medical University (Sechenov University; “Prognosing the efficacy of peptic ulcer therapy on the basis of pharmacogenomics, pharmacometabolomic and pharmacoproteomic technologies of personalized medicine”) for CYP2C19*17 genotyping reagents and consumables.
Disclosure
The authors report no conflicts of interest in this work.
References
Zanger UM, Schwab M. Cytochrome P450 enzymes in drug metabolism: Regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther. 2013;138(1):103–141. | ||
De Morais SM, Wilkinson GR, Blaisdell J, Meyer UA, Nakamura K, Goldstein JA. Identification of a new genetic defect responsible for the polymorphism of (S)-mephenytoin metabolism in Japanese. Mol Pharmacol. 1994;46(4):594–598. | ||
De Morais SM, Wilkinson GR, Blaisdell J, Nakamura K, Meyer UA, Goldstein JA. The major genetic defect responsible for the polymorphism of S-mephenytoin metabolism in humans. J Biol Chem. 1994;269(22):15419–15422. | ||
Sim S, Risinger C, Dahl ML, et al. Common novel CYP2C19 gene variant causes ultrarapid drug metabolism relevant for the drug response to proton pump inhibitors and antidepressants. Clin Pharmacol Ther. 2006;79(1):103–113. | ||
Baldwin RM, Ohlsson S, Pedersen RS, et al. Increased omeprazole metabolism in carriers of the CYP2C19*17 allele; a pharmacokinetic study in healthy volunteers. Br J Clin Pharmacol. 2008;65(5):767–774. | ||
Swen JJ, Nijenhuis M, de Boer A, et al. Pharmacogenetics: from bench to byte-an update of guidelines. Clin Pharmacol Ther. 2011;89(5):662–673. | ||
Balian JD, Sukhova N, Harris JW, et al. The hydroxylation of omeprazole correlates with S-mephenytoin metabolism: a population study. Clin Pharmacol Ther. 1995;57(6):662–669. | ||
Andersson T, Regårdh CG, Lou YC, Zhang Y, Dahl ML, Bertilsson L. Polymorphic hydroxylation of S-mephenytoin and omeprazole metabolism in Caucasian and Chinese subjects. Pharmacogenetics. 1992;2(1):25–31. | ||
Ohnishi A, Murakami S, Akizuki S, Mochizuki J, Echizen H, Takagi I. In vivo metabolic activity of CYP2C19 and CYP3A in relation to CYP2C19 genetic polymorphism in chronic liver disease. J Clin Pharmacol. 2005;45(11):1221–1229. | ||
Scott SA, Sangkuhl K, Stein CM, et al; Clinical Pharmacogenetics Implementation Consortium. Clinical pharmacogenetics implementation consortium guidelines for CYP2C19 genotype and clopidogrel therapy: 2013 update. Clin Pharmacol Ther. 2013;94(3):317–323. | ||
Crettol S, Déglon JJ, Besson J, et al. Methadone enantiomer plasma levels, CYP2B6, CYP2C19, and CYP2C9 genotypes, and response to treatment. Clin Pharmacol Ther. 2005;78(6):593–604. | ||
Yamada H, Dahl ML, Lannfelt L, Viitanen M, Winblad B, Sjöqvist F. CYP2D6 and CYP2C19 genotypes in an elderly Swedish population. Eur J Clin Pharmacol. 1998;54(6):479–481. | ||
Man M, Farmen M, Dumaual C, et al. Genetic variation in metabolizing enzyme and transporter genes: comprehensive assessment in 3 major East Asian subpopulations with comparison to Caucasians and Africans. J Clin Pharmacol. 2010;50(8):929–940. | ||
Sánchez MB, Herranz JL, Leno C, et al. Genetic factors associated with drug-resistance of epilepsy: relevance of stratification by patient age and aetiology of epilepsy. Seizure. 2010;19(2):93–101. | ||
Gaikovitch EA, Cascorbi I, Mrozikiewicz PM, et al. Polymorphisms of drug-metabolizing enzymes CYP2C9, CYP2C19, CYP2D6, CYP1A1, NAT2 and of P-glycoprotein in a Russian population. Eur J Clin Pharmacol. 2003;59(4):303–312. | ||
Payan M, Rouini MR, Tajik N, Ghahremani MH, Tahvilian R. Hydroxylation index of omeprazole in relation to CYP2C19 polymorphism and sex in a healthy Iranian population. Daru. 2014;22(1):81. | ||
Noubarani M, Kobarfard F, Motevalian M, Keyhanfar F. Variation in omeprazole pharmacokinetics in a random Iranian population: a pilot study. Biopharm Drug Dispos. 2012;33(6):324–331. | ||
Tanaka S, Uchida S, Inui N, Takeuchi K, Watanabe H, Namiki N. Simultaneous LC-MS/MS analysis of the plasma concentrations of a cocktail of 5 cytochrome P450 substrate drugs and their metabolites. Biol Pharm Bull. 2014;37(1):18–25. | ||
De Andrés F, Terán S, Bovera M, Fariñas H, Terán E, LLerena A. Multiplex phenotyping for systems medicine: a one-point optimized practical sampling strategy for simultaneous estimation of CYP1A2, CYP2C9, CYP2C19, and CYP2D6 activities using a cocktail approach. OMICS. 2016;20(2):88–96. | ||
Puris E, Pasanen M, Gynther M, et al. A liquid chromatography-tandem mass spectrometry analysis of nine cytochrome P450 probe drugs and their corresponding metabolites in human serum and urine. Anal Bioanal Chem. 2017;409(1):251–268. | ||
Gumus E, Karaca O, Babaoglu MO, et al. Evaluation of lansoprazole as a probe for assessing cytochrome P450 2C19 activity and genotype-phenotype correlation in childhood. Eur J Clin Pharmacol. 2012;68(5):629–636. | ||
Niioka T, Uno T, Sugimoto K, Sugawara K, Hayakari M, Tateishi T. Estimation of CYP2C19 activity by the omeprazole hydroxylation index at a single point in time after intravenous and oral administration. Eur J Clin Pharmacol. 2007;63(11):1031–1038. | ||
Rost KL, Brösicke H, Brockmöller J, Scheffler M, Helge H, Roots I. Increase of cytochrome P450IA2 activity by omeprazole: evidence by the 13C-[N-3-methyl]-caffeine breath test in poor and extensive metabolizers of S-mephenytoin. Clin Pharmacol Ther. 1992;52(2):170–180. | ||
Parkinson A, Hurwitz A. Omeprazole and the induction of human cytochrome P-450: a response to concerns about potential adverse effects. Gastroenterology. 1991;100(4):1157–1164. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.


