Back to Journals » Cancer Management and Research » Volume 10
Urinary exosomal expression of long non-coding RNAs as diagnostic marker in bladder cancer
Authors Yazarlou F, Modarressi MH, Mowla SJ, Kholghi Oskooei V , Motevaseli E, Farhady Tooli L , Nekoohesh L, Eghbali M, Ghafouri-Fard S , Afsharpad M
Received 1 September 2018
Accepted for publication 26 October 2018
Published 26 November 2018 Volume 2018:10 Pages 6357—6365
DOI https://doi.org/10.2147/CMAR.S186108
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Harikrishna Nakshatri
Fatemeh Yazarlou,1 Mohammad Hossein Modarressi,1 Seyed Javad Mowla,2 Vahid Kholghi Oskooei,3 Elahe Motevaseli,4 Leila Farhady Tooli,5 Leila Nekoohesh,6 Maryam Eghbali,1 Soudeh Ghafouri-Fard,3 Mandana Afsharpad7
1Department of Medical Genetics, School of Medicine, Tehran University of Medical Sciences, Tehran, Iran; 2Faculty of Biological Sciences, Department of Genetics, Tarbiat Modares University, Tehran, Iran; 3Department of Medical Genetics, Shahid Beheshti University of Medical Sciences, Tehran, Iran; 4Department of Molecular Medicine, School of Advanced Technologies in Medicine, Tehran University of Medical Sciences, Tehran, Iran; 5Department of Microbiology, School of Biology, College of Science, Tehran University, Tehran, Iran; 6Department of Medical Biotechnology, School of Advanced Technologies in Medicine, Tehran University of Medical Sciences, Tehran, Iran; 7Cancer Control Research Center, Cancer Control Foundation, Iran University of Medical Sciences, Tehran, Iran
Background: Long non-coding RNAs (lncRNAs) and exosomes have been regarded as components of cell signal transmission that modulate indigenous cellular microenvironments. Exosomes also participate in relocation of functional lncRNAs between cells.
Methods: In the present study, we evaluated expression of LINC00355, LINC00958, UCA1-201, UCA1-203, and MALAT1 lncRNAs in urinary exosomes isolated from transitional cell carcinoma (TCC) of bladder, non-malignant urinary disorders, and normal subjects.
Results: LINC00355, UCA1-203, and MALAT1 expression was significantly higher in TCC patients compared to controls (non-malignant or normal samples). However, UCA1-201 expression was significantly decreased in TCC patients compared with controls. LINC00355 and MALAT1 expression was significantly lower in cigarette smokers and opium-addicted TCC patients, respectively. On the other hand, LINC00355 expression tended to be higher in opium-addicted TCC patients. The proposed panel of lncRNAs (composed of UCA1-201, UCA1-203, MALAT1, and LINC00355) had 92% sensitivity and 91.7% specificity for diagnosis of bladder cancer from normal samples.
Conclusion: Transcript levels of lncRNAs in urinary exosomes are potential diagnostic biomarkers in bladder cancer.
Keywords: lncRNA, exosome, bladder cancer
Introduction
Recent reports have highlighted the role of long non-coding RNAs (lncRNAs) in the pathogenesis of several human malignancies including genitourinary cancers.1,2 This underscored part of human transcripts participates in the pathogenesis of transitional cell carcinoma (TCC) of bladder through alteration of cellular pathways of cell transformation.3 In the meantime, as exosomes naturally produce nano-vesicles, which contain proteins and RNAs, they contribute to urothelial carcinogenesis, possibly through alteration of intercellular communications.4 Cooperatively, lncRNAs and exosomes have been regarded as components of cell signal transmission that modulates indigenous cellular microenvironments, since exosomes have role in the relocation of functional lncRNAs between cells.5 Several lncRNAs have been shown to be involved in the pathogenesis of bladder cancer. The metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) upregulation has been detected in bladder cancer samples compared with matched normal urothelium and in high-grade carcinomas compared with low-grade carcinomas. Its roles in enhancement of cell proliferation and motility, while inhibiting apoptosis, have been confirmed in vitro.6 Moreover, MALAT1 has been suggested as an independent prognostic factor in bladder cancer patients and a putative therapeutic target in this kind of malignancy.7 In addition, LINC00958 has an established role in the pathogenesis of bladder cancer, as its silencing decreased cell viability, migration, and invasive potential of these cells. Moreover, LINC00958 has interactions with proteins that are involved in the regulation and initiation of translation and in post-transcriptional modification of RNA and have recognized participation in bladder cancer.8 LINC00355 has been among the most upregulated lncRNAs in bladder cancer samples compared with normal samples as revealed through RNA sequencing of these samples. Moreover, in vitro assessments have demonstrated its contribution in apoptosis, cell proliferation, and migration processes.9 Finally, the urothelial carcinoma-associated (UCA1) lncRNA has been cloned from human bladder TCC cell line. Both in vitro and in vivo assays have confirmed its role in cell proliferation, migration, invasiveness, and drug resistance of TCC cells.10 Several splicing variants have been identified for this lncRNA among which are UCA1-201 (ENST00000397381.5) and UCA1-203 (ENST00000600160.3) with 2,025 bp and 998 bp lengths, respectively. Considering the reported roles of these lncRNAs in the pathogenesis of bladder cancer and the importance of urinary exosomes as diagnostic markers in this malignancy,11 we evaluated expression levels of these lncRNAs in urinary exosomes isolated from TCC patients, normal subjects, and patients with non-malignant urinary disorders to evaluate their diagnostic power.
Patients and methods
Study participants
The study protocol was approved by the Ethical Committee of Tehran University of Medical Sciences. All methods were performed in accordance with the relevant guidelines and regulations. This study was conducted in accordance with the Declaration of Helsinki. All study participants signed the informed consent forms for participation in the study. A total of 59 TCC patients (age [mean ± SD] =61.28±13.01 years), 24 normal subjects (age [mean ± SD] =68±13.56 years), eleven patients with bladder stone (age [mean ± SD] =55.42±155.55 years), six patients with obstructive uropathy (age [mean ± SD] =42.5±8.26 years), and eight patients with benign prostate hyperplasia (age [mean ± SD] =73±7.48 years) participated in the study. All study participants were male. A sample of first morning urine samples was obtained from all participants and centrifuged at 800× g for 10 minutes at 4°C to remove cell pellets and stored at –20°C until further assessments.
Urine exosome isolation
Norgen’s Urine Exosome RNA Isolation Kit (Biotek Corporation, Thorold, ON, Canada) was used for isolation of urine exosomes by spin column method.
Characterization of urine exosomes
Western blotting, dynamic light scattering (DLS) assessments, and electron microscopy were used for assessment of size and morphology of the isolated exosomes.
Western blotting
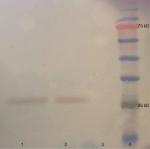
The commercial antibody against exosomal marker protein CD63 was used for probing exosomes after assessment of the protein concentration using Bradford protein assay. BSA was used as the standard sample. Samples were kept at 37°C for 5 minutes and separated on 10% pre-casted gel. Then, they were transferred to nitrocellulose membranes and blocked overnight (5% milk and 0.05% Tween-20 in PBS). Subsequently, they were incubated with primary antibody (Santa Cruz Biotechnology, Dallas, Texas, USA) for 1 hour, washed with PBS, and lastly incubated with secondary horseradish peroxidase-conjugated antibody (SinaClon, Tehran, Iran). The resultant immunoreactive bands were envisioned using chemiluminescent detection system. The prestained protein ladder (SinaClon, Tehran, Iran) was used for assessment of protein size.
Dynamic light scattering assessments
The size of exosomes was also assessed by DLS using a Zetasizer Nano ZS (Malvern Instruments, Malvern, UK) based on the company guidelines.
Electron microscopy
After addition of the slurry b1 solution (Norgen’s Urine Exosome RNA Isolation Kit; Biotek Corporation), a portion of the precipitated exosomes was dissolved in PBS and visualized under scanning electron microscopy (SEM) (QUANTA SEM system; FEI Company, Hillsboro, OR, USA).
Exosomal RNA isolation
Urine Exosome RNA Isolation Kit (Norgen; Biotek Corporation) was used for isolation of RNA from exosomes. This kit uses an all-in-one method for the concentration and isolation of exosomal RNA from urine samples. After binding of the urinary exosomes to a patented resin, RNA is refined from the exosomes using a column-based system.
Quantitative real-time PCR analysis
PrimeScript™ RT reagent Kit (Takara, Tokyo, Japan) was used for cDNA synthesis. Expression levels of lncRNA genes in urine exosomes were measured in the rotor gene 6,000 corbett Real-Time PCR System using RealQ Plus 2x Master Mix Green (Ampliqon, Herlev, Denmark). 5S rRNA was used as normalizer. All experiments were performed in duplicate in 30 µL total volumes. The nucleotide sequence of primers used in expression analyses are shown in Table 1.
  | Table 1 Nucleotide sequence of primers used for expression analysis of lncRNAs in urinary exosomes Abbreviation: lncRNA, long non-coding RNA. |
Statistical methods
All statistical analyses were performed using SPSS v18.0 (SPSS Inc., Chicago, IL, USA). The expression level of each gene was measured by using the following formula: Efficiency^ cycle threshold (CT) normalizer gene/ Efficiency^ CT target gene. The magnitude of expression of each gene between cancerous and control samples was described as fold change value and was calculated by dividing the mean of expression level values in cancerous samples to the matching value of control samples. The differences between relative expressions of genes in defined subcategories of TCC patients were evaluated using independent t-test. Correlation between expression levels of genes in urine samples was assessed using Pearson’s correlation coefficient. The level of significance was set at P<0.05 in all analyses. The receiver operating characteristic (ROC) curve was depicted to assess the suitability of expression levels of each gene or combinations of their expression levels for diagnosis of TCC from normal or non-malignant urinary disorders. The Youden index (j) was applied to obtain the maximum difference between sensitivity (true-positive rate) and 1–specificity (false-positive rate). The diagnostic power of transcript levels was assessed through calculation of the area under curve (AUC) values. AUC values were judged using the following method: 0.90–1= excellent (A), 0.80–0.90= good (B), 0.70–0.80= fair (C), 0.60–0.70= poor (D), and 0.50–0.60= fail (F).
Ethical approval
Ethical approval was obtained from the ethical committee of Tehran University of Medical Science(IR.TUMS.1395.525). Informed consent was obtained from the patients.
Results
General data of patients
The general demographic and clinical data of study participants are summarized in Table 2.
  | Table 2 The general characteristics of study participants Abbreviation: TCC, transitional cell carcinoma. |
Confirmation of exosome size and morphology
Assessment of isolated particles with SEM confirmed that we could effectively isolate exosomes with acceptable quality regarding their size range and morphology (Figure 1). The results were also confirmed by DLS assessment (Figure 2).
  | Figure 2 Size distribution of exosomes by volume as demonstrated by Zetasizer Nano ZS. |
Western blot
We subsequently validated the results of SEM imaging through Western blot assessment of protein lysates acquired from the isolated exosomes. Western blot analysis of samples demonstrated the expression of CD63 as a shared marker of exosomes (Figure 3).
Expression of lncRNA genes in urinary exosomes
We assessed expression of lncRNAs in isolated exosomes of all study participants and compared their expressions between TCC and controls. As demonstrated in Figure 4, there are significant differences in –delta CT values of UCA1-203, UCA1-201, and MALAT1 between TCC patients and total controls.
  | Figure 4 The –delta CT values of lncRNAs in isolated exosomes of TCC patients and controls. Abbreviations: CT, cycle threshold; lncRNAs, long non-coding RNAs; TCC, transitional cell carcinoma. |
LINC00355, UCA1-203, and MALAT1 expression was significantly higher in TCC patients compared to controls (non-malignant urinary disorders or normal samples). However, UCA1-201 expression was significantly decreased in TCC patients compared with controls. Table 3 shows the relative expression of lncRNAs in urinary exosomes of TCC patients compared with controls.
Associations between exosomal expression of lncRNAs and clinicopathological data
LINC00355 and MALAT1 expression was significantly lower in cigarette smokers and opium-addicted TCC patients, respectively. On the other hand, LINC00355 expression tended to be higher in opium-addicted TCC patients. No significant difference was found in expression levels of lncRNAs between different subcategories of TCC patients (Table 4).
Correlations between exosomal expressions of lncRNAs
Significant correlations were found between exosomal expressions of certain lncRNAs in both TCC patients and controls (Table 5). The most significant correlations were found between MALAT1 and LINC00355 in both TCC samples and controls. Moreover, expressions of LINC00985 and UCA1-203 were significantly correlated in TCC samples.
ROC curve analysis
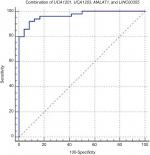
We assessed the diagnostic power of urinary exosome lncRNAs levels in differentiation of TCC from normal samples/total controls (Table 6). Based on the calculated AUC values, UCA1-201 transcript levels had the best performance in differentiation of TCC from normal samples/total controls (AUC values of 0.73 and 0.93, respectively). Of note, the proposed panel of lncRNAs (composed of UCA1-201, UCA1-203, MALAT1, and LINC00355) had 92% sensitivity and 91.7% specificity for diagnosis of bladder cancer from normal samples (Figure 5).
Discussion
Direct contact of cancerous cells with urine makes this bio-fluid a potential source of biomarkers. Urine collection is noninvasive and cost-effective. Different fractions of urine such as urinary sediment, supernatant, and exosome have been assessed to find the most informative biomarkers for genitourinary malignancies.11 Exosomes are nano-vesicles produced by almost all cell types and released into the extracellular microenvironment. These membrane-bound sacs are carriers of major biomolecules including DNAs, mRNAs, miRNAs, lncRNAs, proteins, and lipids which collectively convey a specific message for neighboring or distance cells.4,12 Deciphering the altered messages in certain diseased conditions such as TCC would facilitate recognition of informative diagnostic or prognostic biomarkers. In the current study, we assessed expressions of five lncRNAs in urinary exosomes of TCC patients, urogenital disorders, and normal subjects to appraise their diagnostic power. Of note, we found significant difference in the expression of four lncRNAs including UCA1-201, UCA1-203, MALAT1, and LINC00355 between TCC and normal samples. Bladder cancer-derived exosomes have been previously demonstrated to contain substances that promote angiogenesis and migration of bladder cancer cells and endothelial cells.4 Based on the observed elevated levels of MALAT1 in urinary exosomes of TCC patients compared with normal samples in the current study and the previous reports of participation of MALAT1 in proliferation and motility of bladder cancer cells,6 exosomal transfer of MALAT1 might mediate bladder carcinogenesis. Several previous studies have pointed to the role of UCA1 in the pathogenesis of different cancers and assessed its expression without specification of the transcript variant. For instance, plasma levels of this lncRNA have been suggested as diagnostic markers in gastric13 and bladder cancers.14 UCA1-containing exosomes derived from hypoxic bladder cancer cells have been shown to alter tumor microenvironment in favor of tumor growth and development.14 In the present study using two sets of primers which specifically amplify two separate transcripts, we demonstrated upregulation of UCA1-203 while downregulation of UCA1-201 in TCC samples compared with normal subjects. Such different pattern of expression of these two variants might imply specific roles for them which should be assessed in future studies. Evidence from protein coding genes has shown that alternative splicing can change oncogenic role of a protein to tumor suppressor role.15,16 However, data regarding the presence of such contrary functions of splice variants of lncRNA are scarce due to the significant difference in splicing dynamics between protein coding and lncRNA genes and lower completed splicing index for lncRNAs in the total chromatin fraction.17 Meanwhile, different splice variants of lncRNAs have distinct secondary structure which can define function of the lncRNA isoform.18
Despite the established oncogenic role of LINC00958 in bladder cancer,8 we could not detect any difference in its urinary exosome expression between TCC, normal, and urogenital disorder samples. Such observation might be explained by different expression levels of this lncRNA in secreted exosomes vs cells as has been previously reported for some lncRNAs such as lincRNA-p21, HOTAIR, and ncRNA-CCND1.19
We also demonstrated higher LINC00355 exosomal expression in TCC patients with no history of cigarette smoking compared with smokers and higher MALAT1 expression in TCC patients with no history of opium addiction compared with addicted patients. Few previous reports have highlighted the role of ncRNAs in smoking exposure responses.20 From our observations, one might speculate that opium/cigarette smoking might decrease the level of certain lncRNAs. A previous study hypothesized that smoking might lead to the chromosome damage and deep deletion leading to significant downregulation of some lncRNAs in the context of lung cancer.21 However, another study reported upregulation of lncRNAs in heroin abusers consistent with the anticipated role of lncRNAs as facilitators of extensive alterations in gene expression as happens in drug abuse.22
Pearson’s correlation analyses revealed significant correlations between exosomal expressions of certain lncRNAs in both TCC patients and controls. The most significant correlation was found between MALAT1 and LINC00355 in control samples followed by the correlation between LINC00985 and UCA1-203 in TCC samples. Totally, our data revealed different patterns of correlations between lncRNAs in cancerous and noncancerous samples which might imply dys-regulation of their expression in the context of cancer or a disease-dependent release of lncRNAs in exosomes which warrants further assessments.
We also demonstrated high specificity and sensitivity of the proposed panel of lncRNAs in the diagnosis of bladder cancer. Current diagnosis of bladder cancer relies on cystoscopy which is troublesome for patients and costly for health system.23 Few US Food and Drug Administration-approved urinary biomarkers including protein assays and exfoliated cell tests suffer from imperfect sensitivity and/or specificity values.23 Consequently, establishment of diagnostic or prognostic panels is of critical significance in this malignancy, especially considering the heterogeneous nature and high recurrence rate of this cancer which obligate cystoscopic investigations.24 The current study provides the first clues for application of urinary exosome transcript levels of lncRNAs as diagnostic markers in bladder cancer. The possible application of this panel as prognostic markers needs further studies.
Conclusion
Transcript levels of lncRNAs in urinary exosomes are potential diagnostic biomarkers in bladder cancer.
Consent for publication
The manuscript does not contain any individual person’s data in any form.
Acknowledgments
We thank patients who participated in our study. This article has been extracted from the thesis written by Fatemeh Yazarloo at School of Medicine, Tehran University of Medical Sciences, and is financially supported by a grant from Tehran University of Medical Sciences.
Author contributions
All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
Taheri M, Habibi M, Noroozi R, et al. HOTAIR genetic variants are associated with prostate cancer and benign prostate hyperplasia in an Iranian population. Gene. 2017;613:20–24. | ||
Nikpayam E, Tasharrofi B, Sarrafzadeh S, Ghafouri-Fard S. The role of long non-coding rnas in ovarian cancer. Iran Biomed J. 2017;21(1):3–15. | ||
Wang L, Fu D, Qiu Y, et al. Genome-wide screening and identification of long noncoding RNAs and their interaction with protein coding RNAs in bladder urothelial cell carcinoma. Cancer Lett. 2014;349(1):77–86. | ||
Beckham CJ, Olsen J, Yin PN, et al. Bladder cancer exosomes contain EDIL-3/Del1 and facilitate cancer progression. J Urol. 2014;192(2):583–592. | ||
Hewson C, Morris KV. Form and function of exosome-associated long non-coding RNAs in cancer. Curr Top Microbiol Immunol. 2016;394:41–56. | ||
Han Y, Liu Y, Nie L, Gui Y, Cai Z. Inducing cell proliferation inhibition, apoptosis, and motility reduction by silencing long noncoding ribonucleic acid metastasis-associated lung adenocarcinoma transcript 1 in urothelial carcinoma of the bladder. Urology. 2013;81(1):209.e1–20209. | ||
Li C, Cui Y, Liu LF, et al. High Expression of long noncoding RNA MALAT1 indicates a poor prognosis and promotes clinical progression and metastasis in bladder cancer. Clin Genitourin Cancer. 2017;15(5):570–576. | ||
Seitz AK, Christensen LL, Christensen E, et al. Profiling of long non-coding RNAs identifies LINC00958 and LINC01296 as candidate oncogenes in bladder cancer. Sci Rep. 2017;7(1):395. | ||
Wieczorek E, Reszka E. mRNA, microRNA and lncRNA as novel bladder tumor markers. Clin Chim Acta. 2018;477:141–153. | ||
Wang F, Li X, Xie X, Zhao L, Chen W. UCA1, a non-protein-coding RNA up-regulated in bladder carcinoma and embryo. Influencing Cell Growth and Promoting Invasion. FEBS letters. 2008;582:1919–1927. | ||
Leiblich A. Recent developments in the search for urinary biomarkers in bladder cancer. Curr Urol Rep. 2017;18(12):100. | ||
Théry C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2(8):569–579. | ||
Gao J, Cao R, Mu H. Long non-coding RNA UCA1 may be a novel diagnostic and predictive biomarker in plasma for early gastric cancer. Int J Clin Exp Pathol. 2015;8(10):12936–12942. | ||
Xue M, Chen W, Xiang A, et al. Hypoxic exosomes facilitate bladder tumor growth and development through transferring long non-coding RNA-UCA1. Mol Cancer. 2017;16(1):143. | ||
Silipo M, Gautrey H, Satam S, Lennard T, Tyson-Capper A. How is Herstatin, a tumor suppressor splice variant of the oncogene HER2, regulated? RNA Biol. 2017;14(5):536–543. | ||
Choi JW, Kim DG, Lee AE, et al. Cancer-associated splicing variant of tumor suppressor AIMP2/p38: pathological implication in tumorigenesis. PLoS Genet. 2011;7(3):e1001351. | ||
Tilgner H, Knowles DG, Johnson R, et al. Deep sequencing of subcellular RNA fractions shows splicing to be predominantly co-transcriptional in the human genome but inefficient for lncRNAs. Genome Res. 2012;22(9):1616–1625. | ||
Ward M, McEwan C, Mills JD, Janitz M. Conservation and tissue-specific transcription patterns of long noncoding RNAs. J Hum Transcr. 2015;1(1):2–9. | ||
Gezer U, Özgür E, Cetinkaya M, Isin M, Dalay N. Long non-coding RNAs with low expression levels in cells are enriched in secreted exosomes. Cell Biol Int. 2014;38(9):n/a–1079. | ||
Ehringer MA. Connecting ncRNA cigarette smoking studies with tobacco use behaviors and health outcomes. Front Genet. 2012;3:49. | ||
Liu B, Chen Y, Yang J. LncRNAs are altered in lung squamous cell carcinoma and lung adenocarcinoma. Oncotarget. 2017;8(15):24275–24291. | ||
Michelhaugh SK, Lipovich L, Blythe J, Jia H, Kapatos G, Bannon MJ. Mining Affymetrix microarray data for long non-coding RNAs: altered expression in the nucleus accumbens of heroin abusers. J Neurochem. 2011;116(3):459–466. | ||
Ward DG, Bryan RT. Liquid biopsies for bladder cancer. Transl Androl Urol. 2017;6(2):331–335. | ||
Matullo G, Naccarati A, Pardini B. MicroRNA expression profiling in bladder cancer: the challenge of next-generation sequencing in tissues and biofluids. Int J Cancer. 2016;138(10):2334–2345. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.