Back to Journals » OncoTargets and Therapy » Volume 12
Upregulation of STC2 in colorectal cancer and its clinicopathological significance
Authors Zhang C, Chen S, Ma X, Yang Q, Su F, Shu X, Xie W, Feng M, Xiong B
Received 20 October 2018
Accepted for publication 31 December 2018
Published 15 February 2019 Volume 2019:12 Pages 1249—1258
DOI https://doi.org/10.2147/OTT.S191609
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Takuya Aoki
Chunxiao Zhang,1,* Shuangqian Chen,1,* Xiang Ma,1 Qian Yang,1 Fei Su,1 Xiang Shu,2 Wei Xie,1 Maohui Feng,1 Bin Xiong1
1Department of Gastrointestinal Surgery and Department of Gastric and Colorectal Surgical Oncology, Zhongnan Hospital of Wuhan University, Hubei Key Laboratory of Tumor Biological Behaviors and Hubei Cancer Clinical Study Center, Wuchang District, Wuhan 430071, China; 2Department of Technology, Wuhan Hesheng Medical Technological Company, Wuhan 430071, China
*These authors contributed equally to this work
Background: Stanniocalcin 2 (STC2) is a glycoprotein hormone involved in many biological processes and a secretory protein that regulates malignant tumor progression. The aim of the present study was to further explore the clinicopathological significance and prognostic role of STC2 in colorectal cancer (CRC).
Methods: In this study, STC2 expression was first investigated in Gene Expression Omnibus and The Cancer Genome Atlas, and then validated with the data from our medical center. Univariate and multivariate analyses were performed to assess the association between prognostic factors and survival outcome.
Results: In Gene Expression Omnibus and The Cancer Genome Atlas databases, bioinformatics analysis confirmed that STC2 was significantly increased in CRC compared with that in normal tissues (P<0.01), and CRC patients with high STC2 expression had a shorter overall survival. By analyzing data from our medical center, the results also showed that STC2 expression of CRC tissues was higher than that in normal tissues, whether the transcriptional or protein levels. In the CRC tissues, high STC2 expression was significantly correlated with lymph node metastasis (P=0.047), distant metastasis (P=0.040), and advanced clinical stage (P=0.047). Moreover, Kaplan–Meier analyses indicated that high STC2 expression predicted a worse prognosis, and multivariate Cox regression analysis revealed that STC2 was an independent prognostic factor for overall survival (HR =1.976, 95% CI: 1.092–3.576, P=0.024) in patients with CRC.
Conclusion: Our results suggested that STC2 played an important role in CRC progression and prognosis, and could be a useful biomarker for survival prediction.
Keywords: STC2, prognosis, bioinformatics analysis, colorectal cancer
Introduction
Colorectal cancer (CRC) has been the fourth leading cause of cancer death worldwide for several decades.1 The 5-year survival rate for localized stage CRC is 90%, whereas as CRC spreads to the regional lymph nodes or distant parts of the body, the 5-year survival rate plunges from 71% to 14%.2 Although improvements in treatment protocols, including the discovery of targeted therapies, have substantially improved the clinical outcomes, most CRC patients fail treatment due to local recurrence and distant metastasis.1 Therefore, predicting the risk of distant metastasis and poor prognosis is essential to improve clinical management of CRC. Unfortunately, most of the current medical standard tumor markers lack sensitivity and specificity for the detection of CRC. For the advances of the diagnostic methods, molecular-based diagnostics have been studied and proposed by many groups. Proteomic or genomic approach, based on profiling differentially expressed proteins or genes, has been used for novel biomarker discovery.3–5
We had analyzed differently expressed genes (DEGs) between CRC and paired adjacent normal tissues (PANT) by gene expression microarray and found high expression of the stanniocalcin 2 (STC2) gene in CRC.6 STC2 is a glycoprotein hormone first known to be involved in calcium and phosphate homeostasis.7–9 In addition, STC2 was involved in glutamine or glucose deprivation, and it also had been found to be upregulated under hypoxia, endoplasmic reticulum stress, and radiation.10–13 Microenvironment characterized by hypoxia and low glucose and glutamine supply facilitated the process of solid tumors. STC2 overexpression contributes to tumor cells’ adaptation to such stress conditions, thus promoting tumor progression.10 Recent studies demonstrated that STC2 expression was upregulated in various tumors, including lung cancer,14 breast cancer,15 hepatocellular carcinoma,16 gastric cancer,17 esophageal carcinoma,18 and nasopharyngeal carcinoma.19 Clinical and pathological studies reveal that STC2 overexpression correlates with advanced tumor grade, tumor invasiveness, metastasis, and poor prognosis.16–19 On the other hand, STC2 expression was also upregulated in breast cancer patients; however, the prognosis for these cases was good.20,21 The clinical significance of STC2 in cancer is disputed and depends on cancer type. Furthermore, clinicopathological significance of STC2 gene expression in CRC had been reported in 2009.22 However, its clinical significance and molecular mechanism in carcinogenesis are still not completely understood. Therefore, to further explore the precise role of STC2 for CRC diagnosis and prognosis is essential.
In order to draw a solid conclusion, we first studied STC2 expression in the publicly available Gene Expression Omnibus (GEO) and The Cancer Genome Atlas (TCGA) databases, and then validated it with the data from our medical center. We evaluated the expression of STC2 in several CRC specimens and investigated its association with clinicopathological parameters and overall survival (OS).
Materials and methods
Microarray data processing
The microarray datasets of CRC, including GSE21510, GSE32323, GSE39582, and GSE41328, were downloaded from the public GEO databases (http://www.ncbi.nlm.nih.gov/geo/). Robust Multichip Average, an algorithm used to create an expression matrix from Affymetrix data, was used to adjust the raw files of background. Median levels of transcript expressions were calculated. Gene-level data were then filtered to include only those probe sets with annotations. Probe set annotation mainly reference the new version annotation files that can be download on Affymetrix official website (http://www.affymetrix.com/support/technical/byproduct.affx).
Differentially expressed genes mining
The limma package (http://bioconductor.org/packages/release/bioc/html/limma.html) was used for selection of DEGs.23 The limma package was a correct and popular method for gene selection through differential expression analyses of microarray. Genes that met the cutoff criteria of adjusted P<0.01 and |log2fold change (FC)| ≥2.0 were screened out as DEGs. Among DEGs, STC2 gene was highly expressed in CRC tissues, similar to the result of our microarray data.6 Based on the previous research foundation, the STC2 gene would be further explored.
Validation and survival analysis
To further validate our results, we employed the TCGA database. Expression of the STC2 mRNA in CRC tissues and normal tissues was compared. Kaplan–Meier plotting data of OS of patients with CRC were generated. The patients were classified into high and low STC2 expression groups according to the median STC2 expression level.
Tissue collection
All tissues were acquired from patients who had been diagnosed with CRC by pathological assessment and undergone surgeries at the Zhongnan Hospital of Wuhan University. The study was approved by the Research Ethics Committee of Zhongnan Hospital of Wuhan University. All patients had been adequately informed and written consent was obtained. This was conducted in accordance with the Declaration of Helsinki.
Six CRC tissues and PANT (distance to cancer >5 cm), obtained from patients after surgery between June 2012 and November 2012, made into tissue microarray.6 None of the patients received preoperative radiotherapy or chemotherapy. One-hundred fifteen CRC paraffin-embedded tissues were obtained from the Department of Pathology. The patients received surgical treatment between January 2014 and May 2015 with complete clinicopathological and prognostic information. STC2 protein expression was assessed in these tissues using immunohistochemical staining. Follow-ups were terminated by August 2018.
Immunohistochemical staining
Immunohistochemistry (IHC) of the tissues was performed as described previously.24 Briefly, deparaffinized sections were pretreated with 10 mM sodium citrate buffer for antigen unmasking (pH 6.0, boiling temperature, 30 minutes), blocked in normal serum, incubated with primary antibody of STC2 (1:100, ab63057; Abcam, Cambridge, UK) at 4°C overnight, rinsed, and incubated with secondary antibody (MaxVision™ HRP-Polymer IHC Kit, Maixin-Bio, Fuzhou, China). Signals were amplified using the kit per the manufacturer’s instructions. The positive brown staining was visualized and then photographed using a light microscope at 200× magnification (BX51; Olympus Corporation, Tokyo, Japan).
The IHC evaluation was performed by two independent observers (Xiang Ma and Xiang Shu, the co-authors) who were blinded to the clinical and pathological characteristics associated with the specimens. The extent of STC2 staining was scored by a semiquantitative method in which staining of >10% of the tumor cells was considered positive. The staining intensity was scored as 0 (negative, −), 1 (weak positive, +), 2 (moderated positive, ++), and 3 (strong positive, +++). For analysis, the STC2 protein expression levels were divided into two groups: low expression level (score value <2) and high expression level (score value ≥2).
Western blot
Western blot was completed as described formerly.25 Total protein was extracted with cell lysis buffer (Beyotime, Shanghai, China), and the protein concentration was quantified using an Enhanced BCA Protein Assay Kit (Beyotime). Protein was separated by 10% SDS-PAGE and transferred by semidry electroblotting to polyvinylidene difluoride membranes (Millipore, Billerica, MA, USA). The membrane was blocked for 1 hour with 5% BSA in TBS-T, and probed with corresponding primary antibodies overnight at 4°C, followed by incubation with rabbit or mouse horseradish peroxidase-coupled secondary antibodies for 1 hour. Specific bands were detected using Clarity™ Western ECL Substrate (Bio-Rad, Richmond, CA, USA) on an autoradiographic film. The primary antibodies used were as follows: anti-STC2 (Abcam) and anti-GAPDH (Proteintech, Wuhan, China).
Quantitative real-time polymerase chain reaction
RNA extraction and quantitative real-time polymerase chain reaction (qRT-PCR) were carried out as previously described.25 Total RNA was isolated using RNA plus reagent (Vazyme, Nanjing, China). Complementary DNA was prepared using oligodT primers according to the protocol supplied with the HiScript II Q RT SuperMix for qPCR (Vazyme). Expression of STC2 was determined by qRT-PCR using AceQqPCR SYBR Green Master Mix (Vazyme). Oligonucleotide sequences of the primer sets used were as follows: STC2 forward (5′-TGAAATGTAAGGCCCACGCT-3′) and reverse (5′-CGAGGTGCAGAAGCTCAAGA-3′) and ACTIN forward (5′-CACCCAGCACAATGAAGATCAAGAT-3′) and reverse (5′-CCAGTTTTTAAATCCTGAGTCAAGC-3′).
Statistical analyses
R 3.5.0 was used for microarray analysis. Statistical analyses were performed with the IBM SPSS 22.0 statistical software package (IBM Inc.). The relationship between STC2 expression and clinicopathological features was analyzed using the Chi-square tests. Kaplan–Meier survival curves were constructed, and the log-rank test was carried out using univariate analysis. Multivariate analysis was performed using Cox’s proportional hazards model. P<0.05 was considered statistically significant.
Results
Microarray datasets
The mRNA expression profile datasets, GSE21510, GSE32323, GSE39582, and GSE41328, were generated using the GPL570 microarray platform. One hundred thirty-two samples, consisting of 69 normal tissues and 63 CRC tissues, were employed (Table S1).
Differentially expressed genes
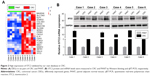
Using R 3.5.0, we identified 1,627, 1,236, 1,607, and 762 DEGs between CRC and normal tissues from the GSE21510, GSE32323, GSE39582, and GSE41328 datasets, respectively (P<0.01, Figures 1A and S1). We found that 381 genes were differentially expressed in all four datasets (Figure 1B). Among the DEGs, STC2 gene was highly expressed in CRC tissue, similar to the result of our microarray data (Figure 2A). Based on the previous research foundation, the STC2 gene underwent further evaluation.6
Validation and survival analyses
To evaluate the expression of STC2 in CRC, we further analyzed STC2 expression in TCGA datasets. The mRNA level of STC2 in the CRC tissues (N=349) and normal tissues (N=41) from TCGA showed that STC2 mRNA expression was significantly upregulated in cancer tissues (P<0.001, Figure 1C). Meanwhile, in 41 paired tissues, the expression of STC2 mRNA was dramatically higher in cancer tissues than that in the PANT (P<0.001, Figure 1D). To further investigate whether the deregulated STC2 correlates with the survivals of the CRC patients, we performed Kaplan–Meier and Cox’s proportional hazards regression model analysis, and found that high STC2 was significantly correlated with poor OS (Figure 1E; Table 1).
STC2 is overexpressed in colorectal cancer
As STC2 showed a consistent pattern of overexpression in GEO and TCGA datasets, our next research direction was to explore the expression pattern of STC2 in the CRC patients of our medical center. In six pairs of specimens, microarray data showed that STC2 gene was significantly upregulated in CRC tissues (Figure 2A). STC2 protein expression was also obviously increased in CRC tissues as determined by Western blot (Figure 2B). Similarly, according to the results of qRT-PCR, STC2 mRNA expression was significantly upregulated in CRC tissues, compared with that in the PANT (Figure 2B).
Associations of STC2 expression with clinicopathological parameters in CRC
The expression of STC2 protein in archived CRC tissue samples was detected by IHC assay. We firstly evaluated STC2 protein expression in 15 pairs of CRC tissues and PANT. STC2 protein was mainly distributed in the cytoplasm and partially in the nucleus. The protein expression of STC2 was significantly increased in CRC compared with that in PANT tissues (Figure 3A).
The association of STC2 expression and clinicopathological features including outcome was analyzed in 115 cases of CRC patients in our medical center. Of the 115 CRC patients, 28 patients (24.35%) had negative STC2 staining (IHC score, 0), 35 patients (30.43%) had weak staining (IHC score, 1), 39 patients (33.91%) had moderated staining (IHC score, 2), and 13 patients (10.43%) had strong staining (IHC score, 3; Figure 3B). Negative and weak stainings were defined as low STC2 expression, whereas moderate and strong stainings were defined as high STC2 expression. The correlations between STC2 expression level and clinicopathological features of CRC were calculated by the chi-squared test and are summarized in Table 2. As shown in Table 2, high STC2 expression in CRC tissues was significantly correlated with lymph node metastasis (P=0.047), distant metastasis (P=0.040), and clinical stage (P=0.047). However, there were no statistically significant relationships between STC2 expression and other clinicopathological variables, such as gender (P=0.595), age (P=0.932), tumor location (P=0.136), tumor size (P=0.105), serum CEA (P=0.904), and tumor infiltration (P=0.205).
High STC2 expression indicated a worse prognosis in colorectal cancer
For all the study subjects, the follow-up period ranged from 1 to 52 months, with a median survival time of 42.8 months. The OS rate was 53.0%, and in patients with high STC2 expression level and those with low level, it was 38.5% and 65.1%, respectively. Additionally, Kaplan–Meier and Cox’s proportional hazards regression model survival analysis revealed that CRC patients with high STC2 expression had a shorter OS (Figure 3C).
To determine whether STC2 expression was an independent prognostic predictor for CRC patients, univariate and multivariate analyses were performed to compare the impact of STC2 expression and other clinicopathological factors on the prognosis. Univariate analysis revealed that clinical variables, including vascular invasion (HR =2.311, 95% CI: 1.302–4.100, P=0.004), lymphatic invasion (HR =1.876, 95% CI: 1.054–3.341, P=0.033), lymph node metastasis (HR =2.391, 95% CI: 1.337–4.275, P=0.003), distant metastasis (HR =5.057, 95% CI: 2.647–9.662, P<0.001), clinical stage (HR =3.015, 95% CI: 1.650–5.508, P<0.001), and STC2 (HR =2.273, 95% CI: 1.278–4.042, P=0.005), were significantly associated with OS of CRC patients (Table 3). Furthermore, in multivariate Cox regression analysis, only distant metastasis (HR =3.742, 95% CI: 1.633–8.575, P=0.002) and STC2 expression (HR =1.976, 95% CI: 1.092–3.576, P=0.024) were independent prognostic factors for CRC (Table 3). The high expression of STC2 had been consistently observed in the CRC patients with poor OS, indicating that STC2 may be functionally important in CRC pathogenesis.
Discussion
CRC local recurrence and distant metastasis leads to the death of 50,260 patients annually in the United States in 2017.1 Therefore, it is critical to distinguish the high risk of recurrence and metastasis to improve the survival of patients with CRC. Based on our previous microarray data, we identified that STC2 could be a potential new biomarker to diagnose and evaluate prognosis of CRC patients in the present study, which represented the association of STC2 expression with clinicopathological features and prognosis in CRC. By analyzing the data from the GEO and TCGA databases, we confirmed that STC2 mRNA level was significantly upregulated in CRC tissues compared with that in normal tissues. The data from our medical center also demonstrated a higher STC2 level in CRC tissue. Similar to the cellular location of STC2 in other types of carcinoma, STC2 protein was located mainly in the cytoplasm and partially in the nucleus. Additionally, STC2 expression was significantly correlated with lymph node metastasis, distant metastasis, and clinical stage. Moreover, high STC2 expression was associated with worse prognosis in patients undergoing surgery for CRC. These findings suggested that STC2 may be involved in CRC progression, and serve as a useful predictor of prognosis.
STC2 belongs to stanniocalcin gene family, which is a family of secreted glycoprotein hormones that was initially discovered in the corpuscles of stannius, an endocrine gland of bony fish.26,27 STC2 and its homologues STC1, are reported to be involved in calcium and phosphate homeostasis, metabolism, reproduction, stress response, and development.27,28 The human STC2 gene encoding 302 amino acid long protein is widely expressed in the kidney, heart, pancreas, and spleen.29 Emerging evidence has revealed abnormal expression of STC2 in tumor tissues, which might be used as a new biomarker to evaluate prognosis of patients with malignant tumors.16–19,30,31 Previous studies have reported that elevated expression of STC2 in gastric cancer tissues was significantly correlated with depth of tumor invasion, lymph node metastasis, venous invasion, and clinical stage, and predicted a worse outcome.17,32 Similarly, aberrant STC2 expression was also correlated with advanced tumor progression in esophageal squamous-cell cancer, hepatocellular carcinoma, nasopharyngeal cancer, lung cancer, and ovarian cancer.14,18,19,33,34 Ieta et al reported that high STC2 expression in CRC tissue was significantly associated with lymph node metastasis, lymphatic invasion, tumor depth, clinical stage, and worse OS.22 Compared with this study, our study employed 596 CRC samples, 132 from GEO, 349 from TCGA, and 115 from our medical center. We accessed more cases from the public database. Furthermore, we first identified STC2 gene was highly expressed in CRC than in normal tissue and high STC2 in cancer tissue was significantly correlated with poor OS by bioinformatics analysis. Subsequently, data from our medical center further confirmed this result. More cases and secondary validation ensured the reliability of our results. Hence, we proved that STC2 may serve as a useful prognosis biomarker in CRC patients.
The impact of STC2 on malignant biological properties has been preliminarily studied. It is reported that knockdown of STC2 in H460 lung cancer cells suppressed growth, migration, invasion, and G0/G1 cell cycle progression, and attenuated the H2O2-induced oxidative stress on cell viability with a subsequent increase in intracellular ROS levels.14 In ovarian cancer, overexpressed STC2 promoted epithelial–mesenchymal transition, as revealed by the increase of N-cadherin/vimentin but a decrease of E-cadherin levels, and enhanced degree of invasiveness under hypoxic condition, mediated by increasing MMP2 and MMP9 expressions and activating ERK1/2 levels.13 Mechanistically, STC2 could trigger the activation of MMP2, Mus81, PI3K/AKT/Snail, and MAPK signaling pathway in neuroblastoma, hepatocellular carcinoma, head and neck squamous cell carcinoma, and cervical cancer.16,35–37 Interestingly, STC2 acts as an oncogene in many kinds of cancers, but it has been reported to be a polite gene in breast cancer. Studies had reported that STC2 overexpression could suppresses breast cancer cell migration and invasion via the PKC/Claudin-1-mediated signaling, and be associated with favorable prognosis, particularly ER-positive breast cancers.21,38 ER-positive breast cancers are usually low-grade malignancies and can be effectively treated with hormonotherapy, which may explain the good prognosis.39,40 Moreover, STC2 could promote growth and migration under hypoxic conditions, and stimulate P-glycoprotein via the PI3K/Akt signaling pathway to influence oxaliplatin resistance in CRC.41,42 There is little research paper for the molecular mechanism of STC2 on CRC behaviors. Therefore, future studies need to elucidate the molecular mechanisms of STC2 in CRC.
Conclusion
Our results revealed that STC2 is an independent prognostic factor for OS in CRC patients. High STC2 expression is strongly correlated with lymph node metastasis, distant metastasis, advanced clinical stage, and worse clinical outcome. Our findings suggested that STC2 could be a potential new biomarker to diagnose and evaluate prognosis of patients with CRC.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81072152, 81770283), Natural Science Foundation of Hubei Province (2015CFA027), Research Foundation of Health and Family Planning Commission of Hubei Province (WJ2015MA010, WJ2017M249), and Clinical Medical Research Center of Peritoneal Cancer of Wuhan (2015060911020462).
Author contributions
CZ and SC contributed equally to this work. CZ, SC, WX, MF, and BX were responsible for conception and design of the study. CZ, SC, QY, and FS did the experiments, data collection, statistical analyses, and writing of the manuscript. XM and XS participated in data collection and provided statistical expertise. All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
Siegel RL, Miller KD, Fedewa SA, et al. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67(3):177–193. | ||
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. | ||
Kulasingam V, Diamandis EP. Strategies for discovering novel cancer biomarkers through utilization of emerging technologies. Nat Clin Pract Oncol. 2008;5(10):588–599. | ||
Makridakis M, Vlahou A. Secretome proteomics for discovery of cancer biomarkers. J Proteomics. 2010;73(12):2291–2305. | ||
Alnabulsi A, Murray GI. Proteomics for early detection of colorectal cancer: recent updates. Expert Rev Proteomics. 2018;15(1):55–63. | ||
Yang Q, Feng M, Ma X, Li H, Xie W. Gene expression profile comparison between colorectal cancer and adjacent normal tissues. Oncol Lett. 2017;14(5):6071–6078. | ||
Flik G, Labedz T, Neelissen JA, Hanssen RG, Bonga SW, Pang PKT. Rainbow trout corpuscles of Stannius: stanniocalcin synthesis in vitro. Am J Physiol Regul Integr Comp Physiol. 1990;258(5):R1157–R1164. | ||
Lu M, Wagner GF, Renfro JL. Stanniocalcin stimulates phosphate reabsorption by flounder renal proximal tubule in primary culture. Am J Physiol Regul Integr Comp Physiol. 1994;267(5):R1356–R1362. | ||
Wagner GF, Jaworski EM, Haddad M. Stanniocalcin in the seawater salmon: structure, function, and regulation. Am J Physiol Regul Integr Comp Physiol. 1998;274(4):R1177–R1185. | ||
Qie S, Liang D, Yin C, et al. Glutamine depletion and glucose depletion trigger growth inhibition via distinctive gene expression reprogramming. Cell Cycle. 2012;11(19):3679–3690. | ||
Ito D, Walker JR, Thompson CS, et al. Characterization of stanniocalcin 2, a novel target of the mammalian unfolded protein response with cytoprotective properties. Mol Cell Biol. 2004;24(21):9456–9469. | ||
Law AYS, Wong CKC. Stanniocalcin-2 is a HIF-1 target gene that promotes cell proliferation in hypoxia. Exp Cell Res. 2010;316(3):466–476. | ||
Law AY, Wong CK. Stanniocalcin-2 promotes epithelial-mesenchymal transition and invasiveness in hypoxic human ovarian cancer cells. Exp Cell Res. 2010;316(20):3425–3434. | ||
Na S, Aldonza MB, Sung HJ. Stanniocalcin-2 (STC2): a potential lung cancer biomarker promotes lung cancer metastasis and progression. Biochem Biophys Acta. 2015;1854(6):668–676. | ||
Bouras T, Southey MC, Chang AC, et al. Stanniocalcin 2 is an estrogen-responsive gene coexpressed with the estrogen receptor in human breast cancer. Cancer Res. 2002;62(5):1289–1295. | ||
Wu F, Li TY, Su SC, et al. STC2 as a novel mediator for Mus81-dependent proliferation and survival in hepatocellular carcinoma. Cancer Lett. 2017;388:177–186. | ||
Yokobori T, Mimori K, Ishii H, et al. Clinical significance of stanniocalcin 2 as a prognostic marker in gastric cancer. Ann Surg Oncol. 2010;17(10):2601–2607. | ||
Kita Y, Mimori K, Iwatsuki M, et al. STC2: a predictive marker for lymph node metastasis in esophageal squamous-cell carcinoma. Ann Surg Oncol. 2011;18(1):261–272. | ||
Lin S, Guo Q, Wen J, et al. Survival analyses correlate stanniocalcin 2 overexpression to poor prognosis of nasopharyngeal carcinomas. J Exp Clin Cancer Res. 2014;33(1):26. | ||
Esseghir S, Kennedy A, Seedhar P, et al. Identification of NTN4, Tra1, and STC2 as prognostic markers in breast cancer in a screen for signal sequence encoding proteins. Clin Cancer Res. 2007;13(11):3164–3173. | ||
Todd JR, Ryall KA, Vyse S, et al. Systematic analysis of tumour cell-extracellular matrix adhesion identifies independent prognostic factors in breast cancer. Oncotarget. 2016;7(39):62939–62953. | ||
Ieta K, Tanaka F, Yokobori T, et al. Clinicopathological significance of stanniocalcin 2 gene expression in colorectal cancer. Int J Cancer. 2009;125(4):926–931. | ||
Ritchie ME, Phipson B, Wu D, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. | ||
Tong Z, Yang XO, Yan H, et al. A protective role by interleukin-17F in colon tumorigenesis. PLoS One. 2012;7(4):e34959. | ||
Sun M, Song H, Wang S, et al. Integrated analysis identifies microRNA-195 as a suppressor of Hippo-YAP pathway in colorectal cancer. J Hematol Oncol. 2017;10(1):79. | ||
Sterba T, Wagner GF, Schroedter IC, Friesen HG. In situ detection and distribution of stanniocalcin mRNA in the corpuscles of Stannius of sockeye salmon, Oncorhynchus nerka. Mol Cell Endocrinol. 1993;90(2):179–185. | ||
Yeung BH, Law AY, Wong CK. Evolution and roles of stanniocalcin. Mol Cell Endocrinol. 2012;349(2):272–280. | ||
Honda S, Kashiwagi M, Ookata K, Tojo A, Hirose S. Regulation by 1α,25-dihydroxyvitamin D3 of expression of stanniocalcin messages in the rat kidney and ovary. FEBS Lett. 1999;459(1):119–122. | ||
Chang AC, Jellinek DA, Reddel RR. Mammalian stanniocalcins and cancer. Endocr Relat Cancer. 2003;10(3):359–373. | ||
Zhou H, Li YY, Zhang WQ, Lin D, Zhang WM, Dong WD. Expression of stanniocalcin-1 and stanniocalcin-2 in laryngeal squamous cell carcinoma and correlations with clinical and pathological parameters. PLoS One. 2014;9(4):e95466. | ||
Shen XJ, Gu K, Shi JP, Yao JQ, Wu JC. Increased expression of stanniocalcin 2 is associated with tumor progression after radiotherapy in patients with cervical carcinoma. Int J Clin Exp Pathol. 2014;7(12):8770. | ||
Arigami T, Uenosono Y, Ishigami S, et al. Clinical significance of stanniocalcin 2 expression as a predictor of tumor progression in gastric cancer. Oncol Rep. 2013;30(6):2838–2844. | ||
Wang H, Wu K, Sun Y, et al. STC2 is upregulated in hepatocellular carcinoma and promotes cell proliferation and migration in vitro. BMB Rep. 2012;45(11):629–634. | ||
Wu J, Lai M, Shao C, Wang J, Wei JJ. STC2 overexpression mediated by HMGA2 is a biomarker for aggressiveness of high-grade serous ovarian cancer. Oncol Rep. 2015;34(3):1494–1502. | ||
Volland S, Kugler W, Schweigerer L, Wilting J, Becker J. Stanniocalcin 2 promotes invasion and is associated with metastatic stages in neuroblastoma. Int J Cancer. 2009;125(9):2049–2057. | ||
Yang S, Ji Q, Chang B, et al. STC2 promotes head and neck squamous cell carcinoma metastasis through modulating the PI3K/AKT/Snail signaling. Oncotarget. 2017;8(4):5976–5991. | ||
Wang Y, Gao Y, Cheng H, Yang G, Tan W. Stanniocalcin 2 promotes cell proliferation and cisplatin resistance in cervical cancer. Biochem Biophys Res Commun. 2015;466(3):362–368. | ||
Hou J, Wang Z, Xu H, et al. Stanniocalcin 2 suppresses breast cancer cell migration and invasion via the PKC/claudin-1-mediated signaling. PLoS One. 2015;10(4):e0122179. | ||
Kaufmann M, Morrow M, von Minckwitz G, Harris JR. Biedenkopf Expert Panel Members. Locoregional treatment of primary breast cancer: consensus recommendations from an international expert panel. Cancer. 2010;116(5):1184–1191. | ||
Lee S, Park IH, Park S, et al. Meeting highlights: the second consensus conference for breast cancer treatment in Korea. J Breast Cancer. 2017;20(3):228–233. | ||
Miyazaki S, Kikuchi H, Iino I, et al. Anti-VEGF antibody therapy induces tumor hypoxia and stanniocalcin 2 expression and potentiates growth of human colon cancer xenografts. Int J Cancer. 2014;135(2):295–307. | ||
Yuan Q, Zhan L, Zhang LL, et al. Stanniocalcin 2 induces oxaliplatin resistance in colorectal cancer cells by upregulating P-glycoprotein. Can J Physiol Pharmacol. 2016;94(9):929–935. |
Supplementary materials
  | Table S1 Microarray datasets of mRNA expression profiles |
References
Tsukamoto S, Ishikawa T, Iida S, et al. Clinical significance of osteoprotegerin expression in human colorectal cancer. Clin Cancer Res. 2011;17(8):2444–2450. | ||
Khamas A, Ishikawa T, Shimokawa K, et al. Screening for epigenetically masked genes in colorectal cancer Using 5-Aza-2′-deoxycytidine, microarray and gene expression profile. Cancer Genom Proteom. 2012;9(2):67–75. | ||
Marisa L, Reyniès AD, Duval A, et al. Gene expression classification of colon cancer into molecular subtypes: characterization, validation, and prognostic value. PLoS Med. 2013;10(5):e1001453. | ||
Lin G, He X, Ji H, et al. Reproducibility probability score – incorporating measurement variability across laboratories for gene selection. Nat Biotechnol. 2006;24(12):1476–1477. |
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.