Back to Journals » Cancer Management and Research » Volume 12
Upregulation of Piezo1 Is a Novel Prognostic Indicator in Glioma Patients
Received 28 February 2020
Accepted for publication 1 May 2020
Published 18 May 2020 Volume 2020:12 Pages 3527—3536
DOI https://doi.org/10.2147/CMAR.S251776
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Eileen O'Reilly
Shanqiang Qu,1,* Shuting Li,2,* Zhicheng Hu3
1Department of Neurosurgery, The First Affiliated Hospital, Sun Yat-sen University, Guangzhou 510080, People’s Republic of China; 2Department of Plastic Surgery, The First Affiliated Hospital, Sun Yat-sen University, Guangzhou 510080, People’s Republic of China; 3Department of Burn Surgery, The First Affiliated Hospital, Sun Yat-sen University, Guangzhou 510080, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Zhicheng Hu
Department of Burn Surgery, The First Affiliated Hospital, Sun Yat-sen University, 58 Zhongshan Road II, Guangzhou 510080, People’s Republic of China
Tel +86-20-87755766-8235
Fax +86-20-87755766-8276
Email [email protected]
Shanqiang Qu
Department of Neurosurgery, The First Affiliated Hospital, Sun Yat-sen University, 58 Zhongshan Road II, Guangzhou 510080, People’s Republic of China
Tel +86-20-87755766-8215
Fax +86-20-87755766-8276
Email [email protected]
Purpose: Previous study reported that Piezo1 was highly expressed in glioma and promoted the proliferation of glioma cells, suggesting that Piezo1 overexpression might contribute to the poor prognosis of patients. Thus, this study aimed to identify whether Piezo1 may become a new prognostic biomarker for glioma patients.
Patients and Methods: Firstly, Piezo1 expression of gliomas was analyzed through GEO and Oncomine dataset, and verified by qRT-PCR and immunohistochemistry (IHC) methods. A total of 183 glioma patients were included in this study between January 2010 and December 2014. Kaplan-Meier survival analyses, Cox regression analyses and ROC curve analyses were performed to assess the diagnostic and prognostic values of Piezo1 in glioma patients.
Results: In this study, Piezo1 was identified to be highly expressed in gliomas, and increased with WHO grade. Chi-square test results showed that Piezo1 expression was significantly related to age (P=0.00), WHO grade (P=0.00), Histopathology (P=0.00), IDH1 mutation (P=0.00) and chemotherapy (P=0.00). Kaplan-Meier analysis showed that the overall survival (OS) of patients with high Piezo1 expression was significantly worse than that of patients with low Piezo1 expression (HR=3.39, 95% CI=2.40– 4.81, P< 0.0001). A multivariate Cox regression analysis revealed that Piezo1 might be an independent prognostic factor for glioma patients (HR=1.34, 95% CI=1.23– 1.47, P=0.000). The area under the ROC curve (AUC) of 1-, 3-, and 5-year overall survival for Piezo1 overexpression was 0.820 (P=0.000), 0.849 (P=0.000), and 0.861 (P=0.000), respectively.
Conclusion: Piezo1 was overexpressed in glioma samples. Piezo1 overexpression as an independent prognostic factor adversely affects the prognosis of patients, which could be a new novel prognostic indicator in glioma patients.
Keywords: novel prognostic indicator, Piezo1, brain glioma
Introduction
Human glioma is the most common primary intracranial tumor and is associated with high levels of morbidity and mortality.1 According to the statistical report of the Central Brain Tumor Registry of the United States, the glioma patients in the United States accounted for 26% of all intracranial tumor patients and 81% of all intracranial malignant tumor patients in 2011–2015.2 Although the level of diagnosis and treatment of gliomas has been improved to some extent, the prognosis of high-grade gliomas is still poor. The median overall survival (OS) times for patients with astrocytoma, mixed glioma, and oligodendroglioma are 5.2, 5.6, and 7.2 years, respectively,3 whereas the median OS of patients with glioblastoma multiforme (GBM) is about 15 months.4 The 2-year survival rate of GBM is about 18% to 28%.5 Gliomas can usually be classified in World Health Organization (WHO) I-IV.6 According to current WHO guidelines of 2016, isocitrate dehydrogenase (IDH) mutation and 1p/19q co-deletion have been used as common clinical prognostic biomarkers. Telomerase reverse transcriptase (TERT) promoter mutation often predicts poor prognosis in patients with GBM.7 With the discovery of many more molecular biomarkers,8,9 treatment of glioma can become increasingly individualized. However, the currently used molecular biomarkers still have some limitations that the WHOI grade glioma and GBM have few IDH mutation and 1p/19q co-deletions. Therefore, it is urgent to clarify the biological characteristics of gliomas and find new prognostic markers to improve the level of diagnosis and treatment of patients.
Previously, in addition to genetic mutations, aberrant expression levels of oncogenes have been proposed as markers for risk stratification. Piezo1 is a novel mechanically activated ion channel that can convert physical stimuli such as touch or blood flow into chemical signals.10
In 2018, Chen et al have found that Piezo1, as a mechanically sensitive cation channel, is highly expressed in gliomas and affects the aggression of glioma cells, which may be related to poor prognosis.11 In addition, more and more studies have found that Piezo1 is also involved in some other biological processes such as tumor cell cycle and angiogenesis in other tumor diseases.12,13 However, its prognostic value in clinical glioma patients is still unclear, and whether Piezo1 can act as a molecular marker for the prognosis of glioma patients is still worthy of further study.
Therefore, in this study, we investigated the potential correlations between Piezo1 expression and the clinicopathological features of glioma patients, and identify that whether Piezo1 could become a new prognostic biomarker for glioma patients.
Patients and Methods
Gene Expression Profiles
Piezo1 expression datasets were screened from the GEO14 (https://www.ncbi.nlm.nih. gov/geo/) and Oncomine database (https://www.oncomine.org/resource/login.html). We collected GSE16011 (8 cases of normal brain tissue, 276 cases of tumor), GSE90598 (7 cases of normal brain tissue, 2 strains of normal brain cell line and 16 cases of GBM), the Rickman Brain15 (normal tissue: 6 cases and astrocytoma: 45 cases) and the Bredel Brain216 (normal tissue: 4 cases and GBM: 26 cases) datasets.
Patients and Data
This study was approved by the Ethics Committee of the First Affiliated Hospital of Sun Yat-Sen University, and all patients signed the written informed consent.
This study included 183 glioma patients who underwent surgical resection in our single center between January 2010 and December 2014. In each case, the diagnosis of glioma was confirmed by two independent pathologists, and the clinicopathological parameters were extracted independently by two researchers. Valid follow-up data were available for all patients. OS was defined as the time from diagnosis to the date of death or the date last known alive. In addition, we investigated the IDH1 status of gliomas by immunohistochemistry (IHC).
Cell Lines and Cell Culture
Two human glioma cell lines (U251 and SW1783) were obtained from the Neurosurgery Department of the First Affiliated Hospital of Sun Yat-Sen University, which the use of these cell lines was approved by ethics committee of the First Affiliated Hospital of Sun Yat-Sen University. All cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM; Life Technologies, Carlsbad, CA, USA) supplemented with 10% FBS (Life Technologies) at 37°C with 5% CO2.
Quantitative Real-Time Reverse Transcription (qRT-PCR)
The total RNA was extracted from the 12 paired glioma (the tissue within 2 cm of glioma margin was defined as adjacent tissue) and normal tissue samples (the tissue outside the 3 cm of the glioma margin is defined as normal tissue), which come from the Department of Neurosurgery of our hospital, using Trizol reagent (Invitrogen), and the RNA from each was reverse transcribed using a ReverTra Ace qPCR RT kit (Toyobo, Japan). The primer sequences for Piezo1 were as follows: Piezo1-F: 5ʹ-GGACTCTCGCTGGTCTACCT-3ʹ and Piezo1-R: 5ʹGGGCACAATATGCAGGCAGA-3ʹ. We used the Biosystems SYBR Green Master Mix (Toyobo, Japan) to monitor the reverse transcribed qRT-PCR analysis. Each experiment was repeated three times, and the presented data are the means of three values. The mRNA levels of Piezo1 compared to β-tubulin were measured by qPCR using the 2−ΔΔC(T) method.17
Immunohistochemistry
For analysis of Piezo1 expression, formalin-fixed, paraffin-embedded slides of glioma tissues from 183 patients were examined by IHC. The immunostaining procedures were performed as described previously.12 Briefly, the slides were incubated with primary Piezo1 antibody (cat. no. 15939-1-AP; dilution, 1:200; ProteinTech Group). Piezo1 expression levels were evaluated based on both the cell staining intensity (scored as: 0, unstained; 1, weakly stained; 2, moderately stained; and 3, strongly stained) and the proportion of positively stained cells (scored as: 0, <5%; 1, 6–25%; 2, 26–50%; and 3, >50%).18 To facilitate the statistical analysis, the cell staining intensity and the proportion of positively stained cells were transformed into an immunoreactivity score (IRS) using the formula: IRS = cell staining intensity × positive cell proportion.
The IDH1 status of glioma samples was also determined by IHC. The primary antibody (cat. no. M7001; dilution, 1:50) was purchased from DAKO. IDH1 status was defined as positive if the staining reaction was strong with a clear cytoplasmic immune reaction and negative if the staining was weakly diffuse or cells remained unstained.19
Statistical Analysis
Data analysis and figure creation were carried out using SPSS version 23.0, GraphPad Prism 7.0, and R software version 3.6.1. Piezo1 mRNA expression was compared between groups using the Mann–Whitney U-test. For categorical variables, the Chi-square test was used for comparisons between groups. OS was compared using Kaplan–Meier survival curves with the Log-rank test. Univariate and multivariate Cox regression analyses were conducted in SPSS. Variables with a P-value ≤0.2 on univariate analysis were considered significant and further included in the multivariate analysis.20 Receiver operating characteristic (ROC) curve analysis was performed to evaluate the accuracy of Piezo1 expression for predicting glioma patient survival. The criterion for statistical significance was P<0.05 on a two-tailed test.
Results
Expression of Piezo1 in Human Glioma
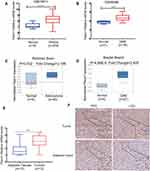
To identify the Piezo1 expression of glioma, we firstly analyzed the mRNA data of GSE16011 dataset (8 samples of normal brain tissue, 276 samples of glioma), GSE90598 dataset (7 samples of normal brain tissue, 2 strains of normal brain cell line and 16 samples of GBM), Rickman Brain dataset (6 normal tissue samples, 45 astrocytoma samples) and Bredel Brain2 dataset (4 normal tissue samples, 27 GBM samples). The statistical results of four datasets showed that, compared with the normal tissue, Pieoz1 expression in glioma was significantly upregulated (P<0.0001, P<0.001, P=0.032 and P<0.001, respectively) (Figure 1A–D).
To verify the above results in the datasets, we further analyze the Piezo1 expression in 12 paired glioma samples (2 paired WHOI, 4 paired WHOII, 3 paired WHOIII and 3 paired WHOIV glioma samples) by the qRT-PCR and IHC method. Consistently, we observed that Piezo1 mRNA levels in glioma were significantly upregulated than that in the adjacent samples (Figure 1E). The IHC results also revealed that the Piezo1 protein levels were overexpressed in glioma compared with that in the adjacent samples (Figure 1F). In addition, the localization of Piezo1 expression in cell lines was detected by immunofluorescence assay, and Piezo1 was found to be predominantly located in the cytoplasm and cell membrane (Supplementary Figure 1)
Characteristics of Glioma Patients
A total of 183 glioma patients were included in this study between January 2010 and December 2014. The ages of all patients ranged from 5 to 79 years, with a mean age of 42.4 years. The median OS for patients was 37.37 months (range, 0.63–119.47 months). Among the 183 cases, 11 cases (6.01%), 73 cases (39.89%), 28 cases (15.30%) and 71 cases (38.80%) were classified as WHO grades I, II, III, and IV of gliomas, respectively. The characteristics of all patients are summarized in Supplementary Table 1.
The Correlation Between Piezo1 Expression and Clinicopathological Features of Glioma
To investigate whether Piezo1 expression (determined by IHC) is correlated with clinicopathological features, 183 patients were divided into high expression group (n=90) and low expression group (n=93) according to the median value of the IRS value of Piezo1. The results of correlations analysis between Piezo1 expression and clinicopathological features are showed in Table 1.
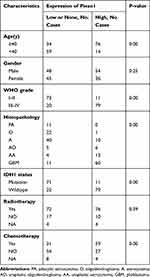 |
Table 1 Correlation Between Piezo1 Expression and Clinicopathologic Characteristics Glioma Patients |
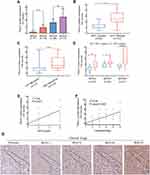
We further analyzed and compared the IRS of Piezo1 in WHOI-IV glioma, and found that Piezo1 expression increased with the increase of glioma WHO grade (Figure 2A), which was confirmed by the IHC results of different clinical stage samples (Figure 2G). Moreover, the IRS of Piezo1 in IDH1-wildtype group was significantly higher than that in IDH1-mutation group (P<0.0001, Figure 2B). The IRS of Piezo1 in high age group (age≥40 years) was significantly higher than that in low age group (age<40 years) (P<0.0001, Figure 2C). We also compared the Piezo1 expression in different histopathological tumors (Supplementary Figure 2). To further assess the positive correlation between Piezo1 expression and IDH1-wildtype, patients were further stratified by WHO grade. The results also showed that the IRS of Piezo1 in IDH1-wildtype group was significantly higher than that in IDH1-mutation group in WHOII-IV subgroup (P<0.01, P<0.01, P<0.0001, Figure 2D).
Through Spearman correlation analysis, we further explored the correlation between Piezo1 expression and WHO grade and histopathology. The results showed the Piezo1 expression was positively related to WHO grade (r=0.74, P<0.0001) and histopathology (r=0.76, P<0.0001), respectively. On linear regression analysis, the correlation between Piezo1 expression levels and WHO grade was linear (R2=0.44, P<0.0001; Figure 2E). The correlation between Piezo1 expression levels and WHO grade was linear (R2=0.44, P<0.0001; Figure 2F).
The Correlation Between Piezo1 Expression and Prognosis of Patients
To explore prognostic value of Piezo1 expression in gliomas, Kaplan-Meier analysis was performed to compare OS of high Piezo1 expression group (n=90) and low Piezo1 expression group (n=93). The results showed that the OS of patients with high Piezo1 expression was significantly worse than that of patients with low Piezo1 expression (HR=3.39, 95% CI=2.40–4.81, P < 0.0001, Figure 3).
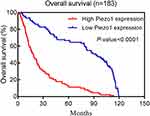 |
Figure 3 Kaplan–Meier postoperative survival curve for patterns of patients with glioma and Piezo1 expression. |
Furthermore, all patients were further stratified by age, gender, WHO grade and IDH1 mutation. The results of subgroup analysis also showed that the OS of patients with high Piezo1 expression was significantly worse than that of patients with low Piezo1 expression (Supplementary Figure 3A–H). Therefore, our results demonstrated that high Piezo1 overexpression could predict a poor prognosis in glioma patients.
Univariate and Multivariate Cox Regression Analysis of Overall Survival in Glioma Patients
To verify whether the impact of Piezo1 overexpression on the OS of glioma patients was independent, we performed univariate and multivariate Cox regression analysis. The results of multivariate analysis showed that Piezo1 overexpression was identified to be an independent prognostic factor for OS in the glioma patients (HR=1.34, 95% CI=1.23–1.47, P=0.000, respectively; Table 2). These results indicated that Piezo1 may be useful as a prognostic biomarker for glioma patients.
 |
Table 2 Univariate and Multivariate Cox Regression Analysis of Prognosis in Patients with Glioma |
Comparison of Piezo1 Overexpression with Existing WHO Grade for Gliomas
To assess the performance of Piezo1 overexpression for predictive the prognosis of glioma patients, a time-dependent ROC analysis was performed. As shown in Figure 4A, the area under the ROC curve (AUC) of 1-, 3-, and 5-year OS for Piezo1 overexpression was 0.820 (P=0.000), 0.849 (P=0.000), and 0.861 (P=0.000), respectively. In addition, the AUC of 1-, 3-, and 5-year OS for WHO grade were 0.769 (P=0.000), 0.833 (P=0.000), and 0.841 (P=0.000), respectively (Figure 4B). Of note, the AUC for the combination of Piezo1 expression and WHO grade to assess the patients’ prognosis was higher (Figure 4C).
 |
Figure 4 ROC curves for the 1-, 3-, and 5-year survival according to the (A) Piezo1 expression, (B) WHO grade and (C) the combination of Piezo1 expression and WHO grade in the glioma (n=183). |
Discussion
The Piezo gene family, including Piezo1 and Piezo2, has been found to encode the molecular components necessary for mechanically sensitive cation channels in mammals.21,22 Piezo1 converts mechanical stimuli into chemical signals to modulate a variety of biological activities, such as sensing blood flow shear stress to guide the correct formation of blood vessels.23 Recently, Piezo1 overexpression has been identified in various tumors.24 In this study, we observed that the Piezo1 expression was significantly upregulated in glioma tissues.
Previous studies have reported that Piezo1 is overexpressed in a variety of solid tumors, including bladder carcinoma, gastric cancer, and prostate cancer.12,25,26 In prostate cancer, Han et al found that Piezo1 overexpression is closely associated with a highly invasive behavior, and that Piezo1 can activate the Akt/mammalian target of rapamycin (mTOR) pathway to contribute to tumorigenesis.12 Moreover, downregulation of Piezo1 expression can significantly inhibit Ca2+ influx, and in turn, inhibit Akt and mTOR phosphorylation, which plays an important role in tumorigenesis and development.27,28 Moreover, in glioma cells, Piezo1 also participates in integrin focal adhesion kinase (FAK) signaling, regulates extracellular matrix production, and reinforces tissue stiffening.11 In turn, stiffening can also activate the opening of Piezo1 channels and further result in Ca2+ influx.11,29 Ca2+ signaling affects the activation of FAK activation, a central molecule in intracellular and extracellular signal transduction, and can promote the progression of cancer.30 In addition, Yang et al observed that interaction between Piezo1 and Trefoil factor family 1 (TFF1) can promote the migration of gastric cancer cells.25 In summary, Piezo1 is upregulated and may act as an oncogene in multiple solid tumors. Our observations are in agreement with those of previous studies. Here, we observed that Piezo1 expression was significantly positively correlated with the WHO grade of gliomas, which showed that Piezo1 upregulation likely plays a positive role in the progression of gliomas. However, interestingly, Huang et al observed significant downregulation of Piezo1 in non-small cell lung cancer tissues31 and found that Piezo1 functional blockade or knock out promotes tumor formation, which suggests that Piezo1 could act as a tumor suppressor gene in NSCLC.31 These findings are in contrast with most studies that support a role of Piezo1 in the development of gliomas. Thus, Piezo1 is associated with a variety of malignant behaviors of tumor cells and plays different roles in different types of tumors.
Furthermore, our results showed that Piezo1 overexpression was negatively correlated with the OS of patients, and was an independent prognostic factor. This observation is consistent with the results obtained in mouse models in which implantation of G532 or G411 cell lines with constitutive knockdown of Piezo1 resulted in a better prognosis than implantation of cell lines with unaltered Piezo1 expression.11 Another reason for the poor prognosis may be the high invasive ability of tumor cells in patients with high Piezo1 expression. Tumor cells with high Piezo1 expression have been shown to have enhanced invasive ability.12 Complete resection of highly invasive gliomas is difficult, and such cases have a high recurrence rate after operation.
Moreover, the result of multivariate analysis suggested that age and WHO grade are also prognostic factors for glioma patients. Age is now widely believed to be an independent risk factor for poor prognosis in gliomas,32 and our results are consistent with those of previous studies.33
There are some limitations to this study. One limitation of this study is the lack of the data of resection extent. The second limitation is that this study focused on the expression and prognostic value of Piezo1 in gliomas.
Conclusions
Piezo1 was significantly upregulated in gliomas. Piezo1 overexpression as an independent prognostic factor adversely affects the prognosis of glioma patients, which could be a new novel prognostic indicator in glioma patients.
Abbreviations
IHC, immunohistochemistry; FFPE, formalin-fixed paraffin-embedded; OS, overall survival; IRS, immunoreactivity score; ROC, receiver operator characteristic curve; TFF1, Trefoil factor family 1; NSCLC, non-small cell lung cancer; TERT, telomerase reverse transcriptase; IDH, isocitrate dehydrogenase.
Data Sharing Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Ethics and Consent Statement
Written informed consent was obtained from each patient. The study protocol was approved by the Medical Ethics Committee of the first affiliated hospital of Sun Yat-Sen University and conducted in accordance with the Declaration of Helsinki.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that they have no conflicts of interest.
References
1. Seidu RA, Wu M, Su Z, et al. Paradoxical role of high mobility group box 1 in glioma: a suppressor or a promoter? Oncol Rev. 2017;11:325. doi:10.4081/oncol.2017.325
2. Ostrom QT, Gittleman H, Truitt G, et al. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2011–2015. Neuro Oncol. 2018;20:iv1–iv86. doi:10.1093/neuonc/noy131
3. Claus EB, Walsh KM, Wiencke JK, et al. Survival and low-grade glioma: the emergence of genetic information. Neurosurg Focus. 2015;38:E6. doi:10.3171/2014.10.focus12367
4. Killock D. Lomustine-temozolomide combination efficacious in newly diagnosed glioblastoma. Nat Rev Clin Oncol. 2019;16:273. doi:10.1038/s41571-019-0192-6
5. Alimohammadi E, Bagheri SR, Taheri S, et al. The impact of extended adjuvant temozolomide in newly diagnosed glioblastoma multiforme: a meta-analysis and systematic review. Oncol Rev. 2020;14:461. doi:10.4081/oncol.2020.461
6. Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131:803–820. doi:10.1007/s00401-016-1545-1
7. Eckel-Passow JE, Lachance DH, Molinaro AM, et al. Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. N Engl J Med. 2015;372:2499–2508. doi:10.1056/NEJMoa1407279
8. Wu C, Su J, Wang X, et al. Overexpression of the phospholipase A2 group V gene in glioma tumors is associated with poor patient prognosis. Cancer Manag Res. 2019;11:3139–3152. doi:10.2147/CMAR.S199207
9. Cai J, Zuo X, Chen Z, et al. Prognostic value and clinical significance of long noncoding RNA CASC2 in human malignancies: a meta-analysis. Cancer Manag Res. 2018;10:1403–1412. doi:10.2147/cmar.s161373
10. Ranade SS, Qiu Z, Woo SH, et al. Piezo1, a mechanically activated ion channel, is required for vascular development in mice. Proc Natl Acad Sci U S A. 2014;111:10347–10352. doi:10.1073/pnas.1409233111
11. Chen X, Wanggou S, Bodalia A, et al. A feedforward mechanism mediated by mechanosensitive ion channel PIEZO1 and tissue mechanics promotes glioma aggression. Neuron. 2018;100:799–815 e797. doi:10.1016/j.neuron.2018.09.046
12. Han Y, Liu C, Zhang D, et al. Mechanosensitive ion channel Piezo1 promotes prostate cancer development through the activation of the Akt/mTOR pathway and acceleration of cell cycle. Int J Oncol. 2019;55:629–644. doi:10.3892/ijo.2019.4839
13. Kang H, Hong Z, Zhong M, et al. Piezo1 mediates angiogenesis through activation of MT1-MMP signaling. Am J Physiol Cell Physiol. 2019;316:C92–C103. doi:10.1152/ajpcell.00346.2018
14. Edgar R, Domrachev M, Lash AE. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi:10.1093/nar/30.1.207
15. Rickman DS, Bobek MP, Misek DE, et al. Distinctive molecular profiles of high-grade and low-grade gliomas based on oligonucleotide microarray analysis. Cancer Res. 2001;61:6885–6891.
16. Bredel M, Bredel C, Juric D, et al. Functional network analysis reveals extended gliomagenesis pathway maps and three novel MYC-interacting genes in human gliomas. Cancer Res. 2005;65:8679–8689. doi:10.1158/0008-5472.can-05-1204
17. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods (San Diego, Calif). 2001;25:402–408. doi:10.1006/meth.2001.1262
18. Guo L, Ding Z, Huang N, et al. Forkhead Box M1 positively regulates UBE2C and protects glioma cells from autophagic death. Cell Cycle. 2017;16:1705–1718. doi:10.1080/15384101.2017.1356507
19. Capper D, Weissert S, Balss J, et al. Characterization of R132H mutation-specific IDH1 antibody binding in brain tumors. Brain Pathol. 2010;20:245–254. doi:10.1111/j.1750-3639.2009.00352.x
20. Atchison CM, Arlikar S, Amankwah E, et al. Development of a new risk score for hospital-associated venous thromboembolism in noncritically ill children: findings from a large single-institutional case-control study. J Pediatr. 2014;165:793–798. doi:10.1016/j.jpeds.2014.05.053
21. Coste B, Mathur J, Schmidt M, et al. Piezo1 and Piezo2 are essential components of distinct mechanically activated cation channels. Science. 2010;330:55–60. doi:10.1126/science.1193270
22. Zhang T, Chi S, Jiang F, et al. A protein interaction mechanism for suppressing the mechanosensitive Piezo channels. Nat Commun. 2017;8:1797. doi:10.1038/s41467-017-01712-z
23. Wang Y, Chi S, Guo H, et al. A lever-like transduction pathway for long-distance chemical- and mechano-gating of the mechanosensitive Piezo1 channel. Nat Commun. 2018;9:1300. doi:10.1038/s41467-018-03570-9
24. Li C, Rezania S, Kammerer S, et al. Piezo1 forms mechanosensitive ion channels in the human MCF-7 breast cancer cell line. Sci Rep. 2015;5:8364. doi:10.1038/srep08364
25. Yang XN, Lu YP, Liu JJ, et al. Piezo1 is as a novel trefoil factor family 1 binding protein that promotes gastric cancer cell mobility in vitro. Digest Dis Sci. 2014;59:1428–1435. doi:10.1007/s10620-014-3044-3
26. Etem EO, Ceylan GG, Ozaydin S, et al. The increased expression of Piezo1 and Piezo2 ion channels in human and mouse bladder carcinoma. Adv Clin Exp Med. 2018;27:1025–1031. doi:10.17219/acem/71080
27. Hirsch FR, Lippman SM. Advances in the biology of lung cancer chemoprevention. J Clin Oncol. 2005;23:3186–3197. doi:10.1200/jco.2005.14.209
28. Dai R, Chen R, Li H. Cross-talk between PI3K/Akt and MEK/ERK pathways mediates endoplasmic reticulum stress-induced cell cycle progression and cell death in human hepatocellular carcinoma cells. Int J Oncol. 2009;34:1749–1757. doi:10.3892/ijo_00000306
29. Friedrich EE, Hong Z, Xiong S, et al. Endothelial cell Piezo1 mediates pressure-induced lung vascular hyperpermeability via disruption of adherens junctions. Proc Natl Acad Sci U S A. 2019;116:12980–12985. doi:10.1073/pnas.1902165116
30. Tai YL, Chen LC, Shen TL. Emerging roles of focal adhesion kinase in cancer. Biomed Res Int. 2015;2015:690690. doi:10.1155/2015/690690
31. Huang Z, Sun Z, Zhang X, et al. Loss of stretch-activated channels, PIEZOs, accelerates non-small cell lung cancer progression and cell migration. Biosci Rep. 2019;39. doi:10.1042/BSR20181679
32. Zhao YY, Chen SH, Hao Z, et al. A nomogram for predicting individual prognosis of patients with low-grade glioma. World Neurosurg. 2019;130:e605–e612. doi:10.1016/j.wneu.2019.06.169
33. Fatehi M, Hunt C, Ma R, et al. Persistent disparities in survival for patients with glioblastoma. World Neurosurg. 2018;120:e511–e516. doi:10.1016/j.wneu.2018.08.114
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.