Back to Journals » Cancer Management and Research » Volume 12
Upregulation of miR-340 Inhibits Tumor Growth and Mesenchymal Transition via Targeting c-MET in Glioblastoma
Authors Lin N, Li W, Wang X, Hou S , Yu D, Zhao X, Jin C, Yao G, Yan W, You Y
Received 21 February 2020
Accepted for publication 22 April 2020
Published 12 May 2020 Volume 2020:12 Pages 3343—3352
DOI https://doi.org/10.2147/CMAR.S250772
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Eileen O'Reilly
Ning Lin,1,* Wentao Li,2,* Xiefeng Wang,2,* Shiqiang Hou,1 Dong Yu,1 Xingyuan Zhao,1 Chunjing Jin,3 Guoquan Yao,1 Wei Yan,2 Yongping You2
1Department of Neurosurgery, Chuzhou Clinical College of Anhui Medical University, The First People’s Hospital Chuzhou, Chuzhou, People’s Republic of China; 2Department of Neurosurgery, The First Affiliated Hospital of Nanjing Medical University, Nanjing, People’s Republic of China; 3Laboratory Medicine Center, Affiliated Hospital of Nantong University, Nantong, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Ning Lin
Department of Neurosurgery, Chuzhou Clinical College of Anhui Medical University, The First People’s Hospital Chuzhou, Chuzhou, People’s Republic of China
Email [email protected]
Yongping You
Department of Neurosurgery, The First Affiliated Hospital of Nanjing Medical University, Nanjing, People’s Republic of China
Email [email protected]
Background: Epithelial–mesenchymal Transition (EMT) is involved in various cancers including glioblastoma. Our previous study has shown that miR-340 negatively correlated with EMT process in glioblastoma.
Purpose: In the present study, we aim to explore the underlying molecular mechanisms of miR-340 in EMT process of glioblastomas.
Materials and Methods: Using RT-qPCR assay, we analyzed the expression of miR-340 in glioma cell lines and normal human glia (NHA) cell line. Using CCK8, Colony formation assays, transwell and Western blot assays, we investigated tumor growth and EMT process. Using luciferase reporter assay, we confirmed a target of miR-340.
Results: Our results showed that miR-340 was down-regulated in glioma cell lines (U87, U251 and LN229) compared to NHA cells. MiR-340 overexpression remarkably inhibited cell proliferation and invasion as well as up-regulated E-cadherin expression and down-regulated N-cadherin, Vimentin, ZEB1, Slug and Snail expressions in U251 and LN229 cells. Further studies have confirmed c-MET as a target gene of miR-340. The EMT-inhibitory effect of miR-340 was lost after c-MET expression was restored. We also identified the antitumorigenic activity of miR-340 in vivo.
Conclusion: These results demonstrated that miR-340 functioned as a tumor suppressor via targeting EMT process and could be a potential therapeutic candidate for treating glioblastomas.
Keywords: miR-340, EMT, c-MET, glioblastoma
Introduction
Glioma is the most common primary intracranial malignant tumor with a poor prognosis.1,2 Glioblastoma (GBM) is the most lethal glioma with a median survival of less than 15 months and often proves intractable to traditional cancer treatments such as surgery, chemotherapy and radiation.3,4 GBM cells have a very strong ability to invade the surrounding normal tissues at the early stage of tumor formation. Therefore, even all tumor tissues are removed, the recurrence at the edge of the surgical cavity cannot be avoided.5–7 Presently, with the development of new generation sequencing technology and the progress of new research methods and novel ideas, the exploration of new treatments for glioblastoma has been the forefront of the efforts of the majority of scientific researchers.
Epithelial–mesenchymal Transition (EMT) refers to the transformation of epithelial cells to mesenchymal stem cells driven by specific physiological or pathological factors.8,9 During the EMT process, epithelial cells lose their polarity and gain deformation and movement ability, further separating from the primary sites and migrating to distant tissues. Meanwhile, the expression of E-cadherin, an adhesion molecule of epithelial cells, decreased, while the expression of mesenchymal cell characteristic molecules (such as Vimentin, N-cadherin, Snail and other mesenchymal proteins) was up-regulated.10,12 To date, EMT process has been found to play an important role in the formation of malignant phenotypes such as invasion and metastasis of tumor cells in various epithelial tumors, including liver cancer, lung cancer and breast cancer.
Small RNA (microRNAs, miRNAs) is a kind of tiny, non-coding, length of 18 to 23 nucleoside RNA, involved in regulating the expression of multiple target genes and multiple cellular processes including cell differentiation, stem cell maintenance, and epithelial–mesenchymal transition.13 The abnormal expression of miRNA represents a common feature and can be caused by different mechanisms such as amplification/deletion, chromosomal rearrangements, and epigenetic regulation in various cancers. MiRNAs show characteristic expression signatures in various cancers and can profoundly affect cancer cell behavior.14,15 Depending on the genes targeted, miNRAs can be used as an oncogene and tumor suppressor. By now, numerous studies have shown that miRNA is closely related to the occurrence and development of glioma.14–16 In our previous study, we have found that EMT was closely associated with malignant progression and clinical outcome via database analysis in glioma. Furthermore, integrated analysis of miRNAs and mRNA profiling revealed an EMT biological process associated with a miRNA profile.16 However, the underlying mechanism of the above miRNA profile in regulating EMT process is not mainly understood. In the present study, we found that miR-340, an EMT process associated miRNA, could reverse the EMT process and inhibit tumor growth in vitro and in vivo via targeting c-MET expression.
Materials and Methods
GBM Samples and Cell Lines
MiRNA and mRNA profiling data of GBM were downloaded from the TCGA data portal (http://cancergenome.nih.gov). Ten primary GBM specimens and ten recurrent samples were obtained from the First Affiliated Hospital of Nanjing Medical University (Nanjing, Jiangsu, China). All tissue samples were collected during surgery, frozen immediately in liquid nitrogen, and stored for total RNA or protein extraction. The human GBM cell lines U87, U251 and LN229 (American TypeCulture Collection, Manassas, VA, USA) were authenticated by American Type Culture Collection using the short tandem repeat genotyping method. NHAs (ScienCell) were grown in astrocyte basal medium supplemented with 2% fetal bovine serum and 1% astrocyte growth supplement (ScienCell). This study was approved by the Ethics Committee of Chuzhou Clinical College of Anhui Medical University, the First People’s Hospital Chuzhou and the First Affiliated Hospital of Nanjing Medical University, and written informed consent was obtained from all patients providing tissue specimens.
Lentiviral Packaging and Stable Cell Line Establishment
The lentiviral packaging kit was purchased from Open Biosystems. Lentivirus-carrying hsa-miR-340 or hsa-miR-negative control (miR-NC) was packaged in human embryonic kidney 293T cells and collected from the supernatant, as instructed by the manufacturer’s manual. Stable cell lines were established by infecting lentivirus into U251 and LN229 GBM cells, followed by puromycin selection.
Cell Proliferation, Invasion Assays and Luciferase Assay
CCK-8 and plate colony formation assays were performed to evaluate the proliferative ability of the cells. Cell invasion ability was evaluated by a Matrigel invasion assay.17
U251 and LN229 cells were seeded into a 24-well plate. After cultured overnight, cells were co-transfected with the wildtype or mutated pMIR-reporter luciferase vector plasmid, and transfected with miR-340 or miR-NC mimics. Luciferase assays were done 48 h after transfection via the Dual-Luciferase Reporter Assay System (Promega, WI, USA).
RNA Extraction and qRT-PCR
RNA was isolated from harvested cells or human tissues with Trizol reagent according to the manufacturer’s instructions (Invitrogen). A stem-loop-specific primer method was used to measure the expression levels of miR-340, as described previously. Expression of U6 was used as an endogenous control. The cDNAs were amplified by qRT-PCR using SYBR Premix Dimer Eraser (Takara) on a 7900HT system, and the fold changes were calculated by relative quantification (2−ΔΔCt).
Protein Extraction, Immunoblotting and Immunohistochemistry (IHC Staining)
Protein extraction and Western blot analysis were performed as previously described.18 Representative images from 2 or 3 independent experiments are shown. The antibodies that were used included c-MET, E-cadherin, N-cadherin, Vimentin, ZEB1, Slug, Snail and GAPDH (Sigma Aldrich).
To detect c-MET expression in glioblastoma, IHC staining was performed on tumor tissue using methods described previously.17 c-MET were scored by an IHC score based on staining intensity and percentage of positive cells within the whole tissue section.
Orthotopic Nude Mouse Models and Treatment
Male BALB/c nude mice (6 weeks old) were used in this study. All procedures involving experimental animals were performed according to a protocol approved by Nanjing Medical University. For establishing intracranial xenografts, 5 × 104 LN229 cells transduced with luciferase lentivirus and stably expressing miR-340 or miR-NC were implanted stereotactically were stereotactically implanted as previously described.19 The bioluminescence imaging was employed to determine the Fluc activity for mice. The study was approved by the Institutional Animal Care and Use Committee of Nanjing Medical University. All operations were performed according to international guidelines concerning the care and treatment of experimental animals.
Statistical Analysis
GSEA (Gene Set Enrichment Analysis) was performed to analyze the correlation of miR-340 and EMT biological process. All experiments were performed 3 times, and the data were analyzed with GraphPad Prism 5. Statistical evaluation for data analysis was determined using the t test. The differences were considered to be statistically significant at P < 0.05.
Results
Expression Pattern and Clinical Significance of miR-340 in GBMs
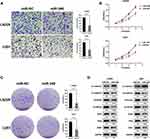
By analyzing the miRNA expression in molecular subtypes of GBM in TCGA database, we determined that the miR-340 was significantly up-regulated in the proneural subtype compared with the other subtypes, especially, the mesenchymal subtype (Figure 1A). GSEA (Gene Set Enrichment Analysis) indicated that EMT process associated genes were more enriched in the GBM patients with low expression level of miR-340 (Figure 1B). Further, the expression of the miR-340 divided patients into two groups with significant different prognosis. Patients with lower expression of miR-340 had significantly shorter overall survival than those with higher miR-340 (p < 0.05) (Figure 1C). It should be highlighted that the expression level of miR-340 was lower in recurrent GBM than those in primary cases (p < 0.05) (Figure 1D). To evaluate the biological role of miR-340 in GBM cells, the expression levels of miR-340 were determined in NHA, U87, U251 and LN229 cell lines. RT-qPCR results showed that miR-340 was significantly down-regulated in GBM cells (U87, U251 and LN229 cell lines) compared to normal glial cell line (NHA) (P < 0.01) (Figure 1E).These data supported the hypothesis that miR-340 are potential tumor suppressors in GBM.
miR-340 Overexpression Inhibited Cell Invasion and Proliferation in vitro
To further determine whether the miR-340 was associated with GBM cell invasion, we restored the expression of miR-340 by ectopic expression of this miRNA in the lowest expression GBM cell line (U251 and LN229) and observed that invasion and proliferation decreased in cells transfected with miR-340 compared with control cells. Transwell assays showed the invasive abilities were significantly decreased (Figure 2A). Furthermore, CCK8 assays and Colony formation assays were performed to assess the role of miR-340 in GBM cell proliferation. The result showed that compared with the control group, forced expression of miR-340 impaired cell proliferative ability in both U251 and LN229 cells (Figure 2B and C). Besides, we also investigated that the effects of miR-340 on EMT process associated markers. As shown in Figure 2D, overexpression of miR-340 resulted lower N-cadherin, Vimentin, ZEB1, Slug, Snail and higher E-cadherin expression. These results indicated that miR-340 overexpression participated in the regulation of GBM cell motility and proliferation in vitro.
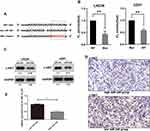
c-MET Is a Target of miR-340 in GBM Cells
To explore the molecular mechanisms through which miR-340 regulates GBM cell proliferation and invasion, we searched candidate target genes of miR-340 using publicly available databases (microRNA.org - Targets and Expression). Among the potential targets, c-MET was chosen for the further analysis (Figure 3A). A luciferase assay was performed in a pMIR-REPORT miRNA reporter vector containing the putative wild-type (WT) and mutant (Mut) c-MET 3′UTR binding sites. Overexpression of miR-340 inhibited wild-type, but not mutant, c-MET reporter activity, suggesting that miR-340 specifically targeted the 3′ UTR of c-MET (Figure 3B). Compared with the stable expression of negative control in U251 and LN229 cells, the stable expression of miR-340 significantly reduced c-MET protein expression (Figure 3C). Furthermore, to explore the relationship between miR-340 and c-MET in clinical specimens, we examined c-MET expression using immunohistochemical analysis on FFPEs of 20 GBM specimens. c-MET expression was down-regulated in the group with high miR-340 levels (Figure 3D and E). Taken together, these results indicated that c-MET was a direct downstream target for miR-340 in GBM cells.
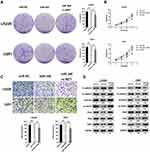
Restoring c-MET Expression Counteracted miR-340’s Suppression on Cell Invasion and EMT Process in GBM Cells
To determine whether miR-340-dependent inhibition of GBM cell proliferation and invasion was indeed mediated by c-MET, we recover the expression of c-MET in U251/miR-340 and LN229/miR-340 cells. Furthermore, we discovered that cell proliferation and invasion were increased (Figure 4A-C). Besides, the downregulation of Mesenchymal Markers-N-cadherin, Vimentin, ZEB1, Slug and Snail, which was thought to be an indirect result of miR-340 overexpression, was rescued by the upregulation of c-MET. Similarly, overexpression of c-MET in miR-340-treated cells down-regulated E-cadherin expression (Figure 4D). In addition, as shown in Supplementary Figure 1, miR-340 inhibited the expression of invasion markers – MMP2 and MMP9. The expression levels of MMP2 and MMP9 by miR-340 were also restored by c-MET overexpression.
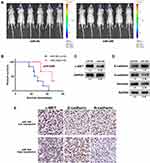
miR-340 Suppressed Glioma Tumor Growth in Orthotopic Models
An in vivo model was established to investigate the relationship between miR-340 and glioma growth. LN229 cells stably expressing miR-340 or miR-NC were injected stereotactically into the brains of nude mice. miR-340-treated LN229 cells displayed a reduction of tumor volume (Figure 5A). To further evaluate the therapeutic effect of miR-340 on nude mice, the survival period of each group was analyzed by a Kaplan–Meier curve. The miR-340 treated group showed a significant improvement in survival compared with the control group until the end of the observation period. Only two mice in the miR-340 treatment group died, whereas no mice were alive in the miR-NC treatment group at this time point (p <0.05) (Figure 5B). The c-MET protein levels were significantly lower in xenografts from miR-340-expressing cells than in those from miR-NC control cells (Figure 5C), confirming that miR-340 overexpression suppressed c-MET expression in vivo. Moreover, the expression levels of N-cadherin and Vimentin, decreased, whereas E-cadherin expression increased in the miR-340 group relative to the miR-NC group (Figure 5D). In addition, EMT associated markers were also analyzed by IHC analysis in intracranial tumors of nude mice (Figure 5E).
Discussion
EMT biological process is the conversion of carcinoma cells from an epithelial to a mesenchymal phenotype that associates with a higher cell motility as well as enhanced chemoresistance and represents a crucial event during cancer progression and dissemination.20 miRNAs show characteristic expression signatures in various cancers and can profoundly regulate various biological processes in cancer cells.13,21 In our previous study, we firstly found that EMT biological process is tightly associated malignant progression and clinical outcome in glioma and identify EMT biological process associated miRNA profile in GBM, which may provide potential novel targets for GBM therapy.16 However, additional studies are required to characterize the roles of the miRNAs involved and to identify further miRNAs as molecular targets of therapy for patients with GBM. Presently, miR-340, a negatively correlated miRNA with EMT process, was chosen for further functional investigation. We found that miR-340 functioned as a tumor suppressor via targeting EMT process in GBM cells.
It has been reported miRNAs play an important role in regulating EMT biological process in various tumors, including glioma.22 Yang et al revealed that miR-598, as a tumor suppressor, negatively regulate DERL1 and EMT to suppress the invasion and migration in NSCLC, thereby putatively serving as a novel therapeutic target for NSCLC clinical treatment.23 Wang et al also found that miR-216b inhibits cell proliferation, migration, invasion, and EMT by targeting the expression of FOXM1 in human NSCLC.24 Furthermore, AC1MMYR2, an inhibitor of dicer-mediated biogenesis of Oncomir miR-21, reverses EMT process and suppresses tumor growth and progression.25 In the present study, GSEA analysis showed that EMT biological process was enriched in GBM samples with miR-340 low expression in TCGA database. miR-340 was down-regulated in glioma cell lines (U87, U251 and LN229) compared to normal human glia (NHA) cell line. miR-340 overexpression remarkably inhibited cell proliferation and invasion as well as up-regulated E-cadherin expression and down-regulated N-cadherin, Vimentin, ZEB1 and Snail expressions in U251 and LN229 cells. These results indicated that miR-340, a negatively correlated with EMT biological process, may inhibit cell viability and EMT process in GBM cells.
The receptor tyrosine kinase c-MET is abundant in various cancers.26,27 c-MET was overexpressed and a strong prognostic marker in high-grade gliomas.28 Besides, c-Met expression is associated with time to recurrence in patients with GBM.29 Emerging evidences have demonstrated that c-MET is involved in crucial parts of glioma cell biology like tumor proliferation, migration, invasion, and angiogenesis, as well as stemness.30,31 Recently, Hu et al found that PLB-1001, a highly selective, efficient, and blood-brain-barrier (BBB)-permeable c-MET kinase inhibitor, demonstrated effective suppression of MET-induced glioma progression in cell lines and xenografts.32 It has been reported that miR-449-5b, miR-144-3p, miR-182, and miR-34a specifically bind the c-MET 3′-UTR region and inhibit c-MET transcription.30 To date, the regulation of miR-340 on the expression of c-MET has not reported. In the present study, we found that miR-340 specifically targeted the 3′ UTR of c-MET by luciferase assay and cell viability and EMT process suppressed by miR-340 was lost after c-MET expression was restored. These results revealed that c-MET were a direct target and mediator of miR-340’s suppression on GBM cells.
In summary, our results show that miR-340 is significantly negatively correlated with EMT biological process in GBM. Furthermore, the current study also demonstrated miR-340 as a tumor suppressor by negatively regulating c-MET and EMT process to suppress the invasion and proliferation in GBM cells, thereby serving as a potential therapeutic approach for the clinical treatment of GBM.
Acknowledgment
Ning Lin, Wentao Li, and Xiefeng Wang are co-first authors for this study.
Author Contributions
All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agreed to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Chen R, Smith-Cohn M, Cohen AL, et al. Glioma subclassifications and their clinical significance. Neurotherapeutics. 2017;14(2):284–297. doi:10.1007/s13311-017-0519-x
2. Gusyatiner O, Hegi ME. Glioma epigenetics: from subclassification to novel treatment options. Semin Cancer Biol. 2018;51:50–58. doi:10.1016/j.semcancer.2017.11.010
3. Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. doi:10.1056/NEJMoa043330
4. Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997–1003. doi:10.1056/NEJMoa043331
5. Perry JR, Belanger K, Mason WP, et al. Phase II trial of continuous dose-intense temozolomide in recurrent malignant glioma: RESCUE study. J Clin Oncol. 2010;28(12):2051–2057. doi:10.1200/JCO.2009.26.5520
6. Seystahl K, Wick W, Weller M. Therapeutic options in recurrent glioblastoma–an update. Crit Rev Oncol Hematol. 2016;99:389–408. doi:10.1016/j.critrevonc.2016.01.018
7. Campos B, Olsen LR, Urup T, et al. A comprehensive profile of recurrent glioblastoma. Oncogene. 2016;35(45):5819–5825. doi:10.1038/onc.2016.85
8. Tiezzi DG, Fernandez SV, Russo J. Epithelial mesenchymal transition during the neoplastic transformation of human breast epithelial cells by estrogen. Int J Oncol. 2007;31(4):823–827.
9. Huang Y, Fernandez SV, Goodwin S, et al. Epithelial to mesenchymal transition in human breast epithelial cells transformed by 17beta-estradiol. Cancer Res. 2007;67(23):11147–11157. doi:10.1158/0008-5472.CAN-07-1371
10. Guarino M, Rubino B, Ballabio G. The role of epithelial-mesenchymal transition in cancer pathology. Pathology. 2007;39(3):305–318. doi:10.1080/00313020701329914
11. Guarino M. Epithelial-mesenchymal transition and tumour invasion. Int J Biochem Cell Biol. 2007;39(12):2153–2160. doi:10.1016/j.biocel.2007.07.011
12. Jiang J, Li X, Yin X, et al. Association of low expression of E-cadherin and beta-catenin with the progression of early stage human squamous cervical cancer. Oncol Lett. 2019;17(6):5729–5739. doi:10.3892/ol.2019.10266
13. Mohr AM, Mott JL. Overview of microRNA biology. Semin Liver Dis. 2015;35(1):3–11. doi:10.1055/s-0034-1397344
14. Wang Y, Wang X, Zhang J, et al. MicroRNAs involved in the EGFR/PTEN/AKT pathway in gliomas. J Neurooncol. 2012;106(2):217–224. doi:10.1007/s11060-011-0679-1
15. Godlewski J, Nowicki MO, Bronisz A, et al. Targeting of the Bmi-1 oncogene/stem cell renewal factor by microRNA-128 inhibits glioma proliferation and self-renewal. Cancer Res. 2008;68(22):9125–9130. doi:10.1158/0008-5472.CAN-08-2629
16. Zhang Y, Zeng A, Liu S, et al. Genome-wide identification of epithelial-mesenchymal transition-associated microRNAs reveals novel targets for glioblastoma therapy. Oncol Lett. 2018;15(5):7625–7630. doi:10.3892/ol.2018.8280
17. Zeng A, Yin J, Li Y, et al. miR-129-5p targets Wnt5a to block PKC/ERK/NF-kappaB and JNK pathways in glioblastoma. Cell Death Dis. 2018;9(3):394. doi:10.1038/s41419-018-0343-1
18. Shi Z, Chen Q, Li C, et al. MiR-124 governs glioma growth and angiogenesis and enhances chemosensitivity by targeting R-Ras and N-Ras. Neuro Oncol. 2014;16(10):1341–1353. doi:10.1093/neuonc/nou084
19. Li S, Zeng A, Hu Q, et al. miR-423-5p contributes to a malignant phenotype and temozolomide chemoresistance in glioblastomas. Neuro Oncol. 2017;19(1):55–65. doi:10.1093/neuonc/now129
20. Groger CJ, Grubinger M, Waldhor T, et al. Meta-analysis of gene expression signatures defining the epithelial to mesenchymal transition during cancer progression. PLoS One. 2012;7(12):e51136. doi:10.1371/journal.pone.0051136
21. Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol. 2018;141(4):1202–1207. doi:10.1016/j.jaci.2017.08.034
22. Wu SG, Chang TH, Liu YN, et al. MicroRNA in lung cancer metastasis. Cancers (Basel). 2019;11:2. doi:10.3390/cancers11020265
23. Yang F, Wei K, Qin Z, et al. MiR-598 suppresses invasion and migration by negative regulation of derlin-1 and epithelial-mesenchymal transition in non-small cell lung cancer. Cell Physiol Biochem. 2018;47(1):245–256. doi:10.1159/000489803
24. Wang L, Wang Y, Du X, et al. MiR-216b suppresses cell proliferation, migration, invasion, and epithelial-mesenchymal transition by regulating FOXM1 expression in human non-small cell lung cancer. Onco Targets Ther. 2019;12:2999–3009. doi:10.2147/OTT.S202523
25. Shi Z, Zhang J, Qian X, et al. AC1MMYR2, an inhibitor of dicer-mediated biogenesis of Oncomir miR-21, reverses epithelial-mesenchymal transition and suppresses tumor growth and progression. Cancer Res. 2013;73(17):5519–5531. doi:10.1158/0008-5472.CAN-13-0280
26. Cataisson C, Michalowski AM, Shibuya K, et al. MET signaling in keratinocytes activates EGFR and initiates squamous carcinogenesis. Sci Signal. 2016;9(433):ra62. doi:10.1126/scisignal.aaf5106
27. Saintigny P, William WN
28. Li MY, Yang P, Liu YW, et al. Low c-Met expression levels are prognostic for and predict the benefits of temozolomide chemotherapy in malignant gliomas. Sci Rep. 2016;6:21141. doi:10.1038/srep21141
29. Liu W, Fu Y, Xu S, et al. c-Met expression is associated with time to recurrence in patients with glioblastoma multiforme. J Clin Neurosci. 2011;18(1):119–121. doi:10.1016/j.jocn.2010.05.010
30. Cheng F, Guo D. MET in glioma: signaling pathways and targeted therapies. J Exp Clin Cancer Res. 2019;38(1):270. doi:10.1186/s13046-019-1269-x
31. Li Y, Li A, Glas M, et al. c-Met signaling induces a reprogramming network and supports the glioblastoma stem-like phenotype. Proc Natl Acad Sci U S A. 2011;108(24):9951–9956. doi:10.1073/pnas.1016912108
32. Hu H, Mu Q, Bao Z, et al. Mutational landscape of secondary glioblastoma guides MET-targeted trial in brain tumor. Cell. 2018;175(6):1665–1678 e1618. doi:10.1016/j.cell.2018.09.038
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.