Back to Journals » OncoTargets and Therapy » Volume 13
Upregulation of MiR-1274a is Correlated with Survival Outcomes and Promotes Cell Proliferation, Migration, and Invasion of Colon Cancer
Authors Ren B, Yang B, Li P, Ge L
Received 15 January 2020
Accepted for publication 10 June 2020
Published 17 July 2020 Volume 2020:13 Pages 6957—6966
DOI https://doi.org/10.2147/OTT.S246160
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr XuYu Yang
Bin Ren,1 Binlin Yang,1 Ping Li,2 Liang Ge1
1Department of Gastrointestinal and Anal Disease Surgery, Affiliated Hospital of Weifang Medical University, Weifang, Shandong 261031, People’s Republic of China; 2Department of Pathology, Weifang Nursing Vocational College, Weifang, Shandong 261053, People’s Republic of China
Correspondence: Liang Ge
Department of Gastrointestinal and Anal Disease Surgery, Affiliated Hospital of Weifang Medical University, No. 2428, Yuhe Road, Weifang, Shandong 261031, People’s Republic of China
Tel +86-0536-3089677
Email [email protected]
Purpose: Colon cancer has become one of the primary causes of cancer-related mortality in recent years. MicroRNAs (miRNAs) play important roles in the regulation of target genes expression. Some of these molecules are aberrantly expressed in colon cancer. The aim of this study was to investigate the potential role of miR-1274a in colon cancer.
Patients and Methods: The expression levels of miR-1274a in colon cancer tissues and cell lines were detected using RT-qPCR. The association between miR-1274a expression and clinical features was analyzed by the χ2 test. Then the Kaplan–Meier method and multivariate Cox regression analysis were used to explore the clinical prognostic significance of miR-1274a in colon cancer. Finally, the effects of miR-1274a on cell growth, migration, and invasion were investigated with the CCK-8 assay, colony formation assay, transwell migration, and invasion assays, respectively.
Results: The expression of miR-1274a was increased in colon cancer tissues and cell lines. The miR-1274a expression was associated with lymph node metastasis, vascular invasion, and TNM stage. Patients with high miR-1274a expression had a poor overall survival time compared with those with low miR-1274a expression. Upregulated miR-1274a promoted cell proliferation, migration, and invasion of colon cancer cells, while inhibition of miR-1274a suppressed these cellular activities by targeting FOXO4.
Conclusion: Our study suggested that miR-1274a might function as an oncogene in human colon cancer and be a potential prognostic biomarker and therapeutic target for the treatment of colon cancer.
Keywords: colon cancer, invasion, miR-1274a, migration, prognosis, growth
Introduction
Colon cancer is a frequent common malignancy of the digestive tract in the colon worldwide, which ranks third among gastrointestinal tumors.1,2 In recent years, with the change of people’s diet structures and lifestyle in China, the incidence and development of colon cancer show an increasing trend.3,4 Currently, surgical resection is still the potentially curative strategy to cure colon cancer.5 Although the survival time of colon cancer patients is higher in early stages, most of the colon cancer patients are diagnosed at advanced stages with localized or distant metastasis, which is associated with low survival time and poor prognosis.6 Although the improved progress in diagnosis and treatment for colon cancer, the improvements in outcomes for advanced cancer is still unsatisfactory. Thus, identifying reliable cancer-related molecular markers associated with the progression of colon cancer is necessary.
MicroRNAs (miRNAs) are a kind of small endogenous non-coding RNAs that play important roles in the regulation of target genes expression by binding to 3ʹ-UTR of their target mRNAs, inhibiting translation or inducing mRNA degradation.7 It has been demonstrated that miRNAs are aberrantly expressed in various diseases and play crucial roles in the development and progression of these diseases, including cancers.8 Increasing studies have indicated that miRNAs participated in various biological behaviors, including development, apoptosis, differentiation, growth, and metastasis.9,10 Moreover, numerous studies revealed that miRNAs have remarkable potential value for diagnosis, prognosis, and therapies of cancers.11,12 Several studies indicated that miRNAs are involved in the progression of colon cancer, such as miR-190b,13 miR-708,14 and miR-298.15 The miR-1274a has an abnormal expression in prostate cancer,16 hepatocellular carcinoma,17 and esophageal squamous cell carcinoma.18 In ovarian cancer patients, low levels of circulating miR-1274a were associated with better survival.19 And in rectal cancer, a comprehensive miRNA expression profiling study identified many aberrantly expressed miRNA in tissues, including miR-1274a.20 Colorectal cancer is usually used as one catalogue and some miRNA expression patterns are similar in rectal cancer and colon cancer while some miRNAs expression pattern is different. Thus, exploring the expression pattern, the clinical significance and the biological functions of miR-1274a in colon cancer are still crucial to explore the mechanism of carcinogenesis of colon cancer.
In the current study, we explored the expression of miR-1274a in colon cancer tissues and human colon cancer cell lines. The association between miR-1274a expression and clinicopathological characteristics of colon cancer patients was also analyzed. In addition, the clinical significance and functional role of miR-1274a were investigated.
Patients and Methods
Patients and Specimens
A total of 117 colon cancer patients diagnosed in the Affiliated Hospital of Weifang Medical University were enrolled in this study from January 2011 to December 2014. Among them, 117 paired fresh tissue specimens and corresponding non-tumor tissue specimens during surgical resection were collected and immediately stored in liquid nitrogen for RNA extraction. All patients who provided specimens did not receive any anti-therapies before surgical resection, such as radiotherapy and chemotherapy. The present study was approved by the Ethics Committee of the Affiliated Hospital of Weifang Medical University and all patients signed informed consent. Patients’ basic clinical information was collected and listed in Table 1. The follow-up overall survival and progression-free survival information were collected 5 years for survival analysis.
 |
Table 1 Association of Clinical Characteristics of Colon Cancer Patients and MiR-1274a Expression |
Cells Culture and Transfection
Human colon cancer cell LoVo, SW480, SW620, SW1116, HCT116, and the human colon mucosal epithelial cell line NCM460 were obtained from the Institute of Biochemistry and Cell Biology of Chinese Academy of Sciences (Shanghai, China). All cells were cultured in DMEM medium (Gibco; Thermo Fisher Scientific, Waltham, MA, USA) containing 10% FBS (Gibco; Thermo Fisher Scientific, Waltham, MA, USA) in an atmosphere containing 5% CO2 at 37°C.
The miR-1274a mimic, mimic negative control (mimic NC), miR-1274a inhibitor, inhibitor NC were purchased from GenePharma Co. Ltd (Shanghai, China). Then transfection was performed using the Lipofectamine 2000 reagent (Invitrogen, Carlsbad, MA, USA) according to the manufacturer’s instructions.
RNA Extraction and Reverse Transcription-Quantitative PCR (RT-qPCR)
RNA was extracted from colon cancer tissue or cell lines by using TRIzol reagent (Invitrogen, Carlsbad, MA, USA) following the manufacturer’s protocol. The RNA extraction concentration and quality were measured with a NanoDrop 1000 spectrophotometer by calculating the ratio of the optical density at 260 nm and 280 nm (A260/A280 in 1.8–2.0 was used for further preparations). cDNA was synthesized from total RNA using the Mir-X miRNA First Strand Synthesis Kit (TaKaRa, Dalian, China). Then RT-qPCR was performed to detect the miR-1274a expression with SYBR Premix Ex Taq II (TaKaRa, Dalian, China) on an ABI Prism 7500 Fast Real-Time PCR system (Applied Biosystems, Foster City, CA, USA). U6 was used to normalize the miR-1274a expression levels. And the relative expression levels of miR-1274a was calculated using the 2-ΔΔCt methods. Primer sequences used for RT-PCR were: miR-1274a, 5ʹ-GCCGAGTTCAGGTCCCTGTT-3ʹ (forward) and 5ʹ- CTCAACTGGTGTCGTGGA −3ʹ(reverse); U6, 5ʹ-GCGCGTCGTGAAGCGTTC-3ʹ (forward) and 5ʹ-GTGCAGGGTCCGAGGT-3ʹ (reverse).
Cell Counting Kit-8 (CCK-8) Assay
Cell proliferative capacities of LoVo and SW620 cells were evaluated with the CCK-8 (Sigma, Louis, MO, USA) assay according to the manufacturer’s instructions. The above colon cancer cells (4 × 103 cells/well) were seeded into 96-well plates and incubated in DMEM medium containing 10% FBS at 37°C with 5% CO2. Then 10 μL CCK-8 solution was added to each well at different time periods (0, 24, 48, 72 h) and incubated for 2 h at 37°C. The absorbance value of each well was measured at 450 nm wavelength using a microplate reader.
Colony Formation Assay
After transfection, all the cells in each group were seeded into each well of 6-well plates and cultured at 37°C in an atmosphere containing 5% CO2. After 14 days, the clones (≥ 50 cells) were washed with PBS, fixed with 4% paraformaldehyde, and stained with 0.1% crystal violet. Then the cell clones were counted and compared.
Transwell Migration and Invasion Assays
The migratory and invasive capacities of colon cancer cells were analyzed by the transwell assay using transwell inserts (8 μm pore size; Corning, NY, USA) according to the protocol of the manufacturer. Transfected cells that resuspended in serum-free medium were seeded in the upper chamber without Matrigel coating for migration assay, while precoated with Matrigel (BD Biosciences, San Jose, CA, USA) for invasion assay. Then, the lower chambers were added culture medium with 10% FBS. After 24 h, the migrated and invaded cells were stained with 1% crystal violet. The number of cells was counted from five random fields under a light microscope (Olympus, Toyko, Japan).
Bioinformatic Analysis and Dual-Luciferase Reporter Assay
The software program miRanda (http://www.microrna.org/microrna/home.do) was used to predict the putative target genes of miR-1274a. The dual-luciferase reporter assay was used to confirm the relationship between miR-1274a and FOXO4. The wild-type and mutated 3ʹ-UTR segments of FOXO4 mRNA containing miR-1274a binding sites were amplified and inserted into the pmirGLO vector (Promega, Madison, WI, USA), as FOXO4 3ʹ-UTR-WT and FOXO4 3ʹ-UTR-Mut, respectively. Then the FOXO4 3ʹ-UTR-WT and FOXO4 3ʹ-UTR-Mut vectors were co-transfected with miR-1274a mimic, mimic NC, miR-1274a inhibitor, and inhibitor NC into LoVo cells using the Lipofectamine 2000 reagent (Invitrogen, Carlsbad, MA, USA) as per the manufacturer’s instructions. After 48 h of transfection, luciferase activities were analyzed using a dual-luciferase reporter assay system (Promega, WI).
Statistical Analysis
All experiments were performed independently at least three times each time. All quantified data were presented as the mean ± SD. The statistical analyses were performed with SPSS statistical software (version 20.0; IBM SPSS, USA) and GraphPad Prism software (version 5.0; GraphPad, USA). Pearson’s χ2 analysis was performed to analyze the relationship between miR-1274a expression and clinical characteristics of colon cancer patients. Overall survival and progression-free survival were analyzed using the Kaplan–Meier method. Differences between two groups were analyzed using Student’s t-test, and differences among groups were analyzed using one-way ANOVA followed by Tukey’s post hoc test. The P values less than 0.05 were considered statistically significant.
Results
MiR-1274a Expression in Colon Cancer Tissues and Cell Lines
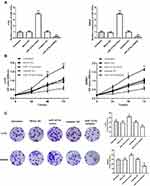
To investigate the role of miR-1274a in colon cancer, we detected miR-1274a expression levels in colon cancer. We found that miR-1274a was highly expressed in colon cancer tissues compared with adjacent non-tumor tissues (P < 0.001, Figure 1A). Then, we tested the miR-1274a expression in cultured colon cancer cell lines and normal colon mucosal epithelial cell line NCM460. As shown in Figure 1B, the expression of miR-1274a levels was higher in colon cancer cells compared to that of the NCM460 cells (P < 0.001). Among these colon cancer cells, LoVo and SW620 cells showed the highest miR-1274a expression levels, which were chosen for subsequently functional analyses.
The Relationship Between MiR-1274a Expression and Clinical Parameters as Well as Prognosis of Colon Cancer Patients
Then we further explored the relationship between miR-1274a expression and clinical characteristics of colon cancer patients. These colon cancer patients were divided into low miR-1274a expression level group and high miR-1274a expression level group according to the median expression level of miR-1274a (1.558) in colon cancer tissues. The χ2 test results in Table 1 showed that the expression of miR-1274a was associated with lymph node metastasis (P = 0.044), vascular invasion (P = 0.046), and TNM stage (P = 0.023). However, there was no significant association between miR-1274a expression and other clinical parameters, such as age, gender, and differentiation (P > 0.05).
Further survival analysis using the Kaplan–Meier method showed that patients with high miR-1274a expression had shorter overall survival time compared with patients with reduced miR-1274a expression (Log rank test, P = 0.013, Figure 2A). In addition, patients with high miR-1274a expression had shorter progression-free survival time (Log rank test, P = 0.019, Figure 2B). Then we used multivariate Cox regression analysis to explore independent prognostic factors for colon cancer. The results in Table 2 showed that the expression of miR-1274a (P = 0.027), lymph node metastasis (P = 0.038), and TNM stage (P = 0.032) were independent prognostic factors for overall survival of colon cancer.
 |
Table 2 Multivariate Cox Analysis of Predictors for Overall Survival of Colon Cancer Patients |
MiR-1274a Overexpression Promotes Cell Growth, Migration, and Invasion of Colon Cancer Cells
To investigate the functional role of miR-1274a in colon cancer, LoVo, and SW620 cells were transfected with miR-1274a mimics or miR-1274a inhibitors, and the transfection efficiency was confirmed by RT-qPCR. As shown in Figure 3A, the expression of miR-1274a was significantly increased by miR-1274a mimics, while that was downregulated by miR-1274a inhibitors, compared with untreated cells (P < 0.001). Then CCK-8 assays and colony formation assays were used to explore the proliferation of colon cancer cells. Both the results showed that overexpression of miR-1274a promoted cell growth capacity, while downregulation of miR-1274a inhibited cell growth capacity of LoVo and SW620 cells, compared with untreated cells (P < 0.05, Figure 3B and C).
In addition, transwell migration and invasion assays were used to explore the migratory and invasive capabilities of LoVo and SW620 cells. Consistent with proliferative results, the results in Figure 4A and B, upregulation of miR-1274a enhanced cell migration and invasion abilities, while knockdown of miR-1274a suppressed cell migration and invasion abilities, compared with untreated cells (P < 0.001).
FOXO4 is a Direct Target of MiR-1274a
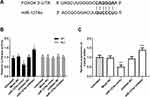
MiR-1274a was identified using miRanda that overlapped many target genes, including FOXO4. As shown in Figure 5A, miR-1274a had putative seed sequences in the 3ʹ-UTR of FOXO4. To evaluate whether FOXO4 may be directly bound by miR-1274a, the dual-luciferase reporter assay was performed. The results showed that FOXO4 3ʹ-UTR-WT was present, transfection of miR-1274a mimics led to a significant decrease in luciferase activity while transfection of miR-1274a inhibitor led to an increase in luciferase activity (P < 0.05, Figure 5B). However, no significant change was observed in the vector containing FOXO4 3ʹ-UTR-Mut. Then we explored the expression levels of FOXO4 mRNA in LoVo cells transfected with miR-1274a mimic or inhibitor. The results in Figure 5C indicated that miR-1274a mimic decreased the expression levels of FOXO4 mRNA, while miR-1274a inhibitor promoted the expression of FOXO4 mRNA (P < 0.001).
Discussion
Metastasis is the leading cause of death for cancer patients. Because the symptoms of early clinical stages of colon cancer patients are not obvious, most patients are initially diagnosed at advanced stages with metastasis.21 Increasing studies demonstrated that abnormal expression of genes was associated with tumorigenesis and progression of cancers, including miRNAs.22,23 For instance, miR-205 was found involved in the angiogenesis and progression of thyroid cancer.24 Numerous studies have also demonstrated that aberrant expression of miRNAs was involved in the development and progression of colon cancer, such as miR-18325 and miR-137.26 Thus, finding more novel cancer-related miRNAs is important to develop specific therapeutic approaches for colon cancer.
In the present study, by comparing adjacent non-tumor tissues, we found that miR-1274a expression was significantly higher in colon cancer tissues. Moreover, we also found that miR-1274a expression was increased in colon cancer cells compared with normal mucosal epithelial cell line NCM460. These results indicated that miR-1274a may play an oncogenic role in colon cancer. The associated of miR-1274a expression with clinical characteristics data were further analyzed. The results showed that the high expression of miR-1274a was associated with positive lymph node metastasis, vascular invasion, and advanced TNM stages, indicating miR-1274a expression may be significantly associated with the occurrence and progression of colon cancer.
Increasing evidence has demonstrated that various miRNAs are related to the survival outcome and prognosis of cancer patients, including colon cancer.27 The miR-1274a expression might play an important role in the progression of ovarian cancer and could be used as a prognostic biomarker for ovarian cancer.19 In breast cancer, miR-1274a was also found significantly associated with overall survival and progression-free survival in metastatic breast cancer patients.28 To explore whether miR-1274a has the potential clinical prognostic significance in colon cancer, we performed Kaplan–Meier curve method with the Log rank test. The results showed that patients with high miR-1274a expression had a shorter overall survival time and progression-free survival time compared with those with low miR-1274a expression. Moreover, multivariate Cox regression assay showed that miR-1274a expression was an independent prognostic predictor for overall survival of colon cancer patients. These results indicated that miR-1274a may be a prognostic biomarker for colon cancer.
Considering miR-1274a expression in colon cancer tissues was associated with lymph node metastasis and TNM stage, we speculate that miR-1274a expression may participate in the growth and metastasis of cancer cells. Thus, we investigated the effects of miR-1274a expression on cell growth, migration, and invasion using colon cancer cells. We found that overexpression of miR-1274a by miR-1274a mimics promoted cell growth, migration, and invasion, conversely, inhibition of miR-1274a by miR-1274a inhibitors suppressed these cellular activities. These results suggested that miR-1274a may be involved in the progression of colon cancer. Previous studies have shown that miR-1274a can regulate different target gene expression, such as BMPR1B,29 ADAM9,17 and FOXO4,30 in different biological processes. For instance, miR-1274a was upregulated in clear cell renal cell carcinoma, and loss of oncogenic miR-1274a reduced cancer cell proliferation and induced apoptosis in clear cell renal cell carcinoma through targeting BMPR1B.29 MiR-1274a is also found upregulated in gastric cancer and promotes gastric cancer cell growth and migration through targeting FOXO4, providing a potential target for the therapy of gastric cancer.30 These previous studies are in concordance with our observed trend for miR-1274a in colon cancer. In the present study, the dual-luciferase reporter assay results confirmed that FOXO4 was a direct target of miR-1274a. Interestingly, FOXO4 was downregulated in colorectal cancer and might act as a tumor suppressor in the development and progression of colorectal cancer.31,32 Thus, we speculate that miR-1274a may also play an oncogenic role in colon cancer and promotes the progression of colon cancer by targeting FOXO4. Considering the potentially crucial role of miR-1274a in colon cancer, the detailed molecular mechanism of miR-1274a will be investigated in future studies.
Conclusion
In conclusion, the expression levels of miR-1274a in colon cancer and para-carcinoma tissues of patients in this study suggest that miR-1274a plays an important role in the development and progression of colon cancer. And the expression of miR-1274a in tissues was associated with the prognosis of colon cancer patients and it might promote cell growth, migration, and invasion by targeting FOXO4. Thus, miR-1274a may be a novel prognostic biomarker, and the miR-1274a/FOXO4 axis may be a therapeutic target for the treatment of colon cancer. The present study may provide novel insight into the improvement of therapeutic strategies for colon cancer patients.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Zhang H, Guo M, Chen H, et al. Effects of CIK on hypoxia inducible factor-1α and T-cell subsets on colon 26 cancer xenograft mice. Oncol Lett. 2016;11:1371–1374. doi:10.3892/ol.2016.4081
2. Zhou W, Pan B, Liu L. Integrated bioinformatics analysis revealing independent prognostic long non-coding RNAs DNAH17-AS1 and RP11-400N13.2 and their potential oncogenic roles in colorectal cancer. Oncol Lett. 2019;18:3705–3715. doi:10.3892/ol.2019.10730
3. Zhang S, Cui Y, Weng Z, et al. Changes on the disease pattern of primary colorectal cancers in Southern China: a retrospective study of 20 years. Int J Colorectal Dis. 2009;24:943–949. doi:10.1007/s00384-009-0726-y
4. Lin S, Jiang T, Yu Y, et al. Secernin-1 contributes to colon cancer progression through enhancing matrix metalloproteinase-2/9 exocytosis. Dis Markers. 2015;2015:230703. doi:10.1155/2015/230703
5. Hermans E, van Schaik PM, Prins HA, et al. Outcome of colonic surgery in elderly patients with colon cancer. J Oncol. 2010;2010:865908. doi:10.1155/2010/865908
6. Ding L, Lan Z, Xiong X, et al. The dual role of microRNAs in colorectal cancer progression. Int J Mol Sci. 2018;19:2791. doi:10.3390/ijms19092791
7. Gao X, Zhao P, Hu J, et al. MicroRNA-194 protects against chronic hepatitis B-related liver damage by promoting hepatocyte growth via ACVR2B. J Cell Mol Med. 2018;22:4534–4544. doi:10.1111/jcmm.13714
8. Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203–222. doi:10.1038/nrd.2016.246
9. Luo J, Pan J, Jin Y, et al. MiR-195-5p inhibits proliferation and induces apoptosis of non-small cell lung cancer cells by targeting CEP55. Onco Targets Ther. 2019;12:11465–11474. doi:10.2147/OTT.S226921
10. Xiang J, Wu Y, Li DS, et al. miR-584 suppresses invasion and cell migration of thyroid carcinoma by regulating the target oncogene ROCK1. Oncol Res Treat. 2015;38:436–440. doi:10.1159/000438967
11. Yang ZQ, Wu CA, Cheng YX. Prognostic value of microRNA-133a expression and its clinicopathologic significance in non-small cell lung cancer: a comprehensive study based on meta-analysis and the TCGA database. Oncol Res Treat. 2018;41:762–768. doi:10.1159/000492343
12. Sun H, Wang L, Zhao Q, et al. Diagnostic and prognostic value of serum miRNA-1290 in human esophageal squamous cell carcinoma. Cancer Biomark. 2019;25:381–387. doi:10.3233/CBM-190007
13. Zhao Q, Liu C, Cui Q, et al. miR-190b promotes colorectal cancer progression through targeting forkhead box protein P2. Exp Ther Med. 2020;19:79–84. doi:10.3892/etm.2019.8175
14. Sun S, Hang T, Zhang B, et al. miRNA-708 functions as a tumor suppressor in colorectal cancer by targeting ZEB1 through Akt/mTOR signaling pathway. Am J Transl Res. 2019;11:5338–5356.
15. Arabsorkhi Z, Gharib E, Yaghmoorian Khojini J, et al. miR-298 plays a pivotal role in colon cancer invasiveness by targeting PTEN. J Cell Physiol. 2020;235:4335–4350.
16. Said R, Garcia-Mayea Y, Trabelsi N, et al. Expression patterns and bioinformatic analysis of miR-1260a and miR-1274a in Prostate Cancer Tunisian patients. Mol Biol Rep. 2018;45:2345–2358. doi:10.1007/s11033-018-4399-x
17. Zhou C, Liu J, Li Y, et al. microRNA-1274a, a modulator of sorafenib induced a disintegrin and metalloproteinase 9 (ADAM9) down-regulation in hepatocellular carcinoma. FEBS Lett. 2011;585:1828–1834. doi:10.1016/j.febslet.2011.04.040
18. Shinozuka E, Miyashita M, Mizuguchi Y, et al. SnoN/SKIL modulates proliferation through control of hsa-miR-720 transcription in esophageal cancer cells. Biochem Biophys Res Commun. 2013;430:101–106. doi:10.1016/j.bbrc.2012.11.005
19. Halvorsen AR, Kristensen G, Embleton A, et al. Evaluation of prognostic and predictive significance of circulating microRNAs in ovarian cancer patients. Dis Markers. 2017;2017:3098542. doi:10.1155/2017/3098542
20. Li X, Zhang G, Luo F, et al. Identification of aberrantly expressed miRNAs in rectal cancer. Oncol Rep. 2012;28:77–84. doi:10.3892/or.2012.1769
21. Liu Z, Li Y, Luo Q. Relationship between CT signs and the expression of miR-146a in colon cancer. Oncol Lett. 2018;16:6598–6602. doi:10.3892/ol.2018.9415
22. Martens-Uzunova ES, Olvedy M, Jenster G. Beyond microRNA–novel RNAs derived from small non-coding RNA and their implication in cancer. Cancer Lett. 2013;340:201–211. doi:10.1016/j.canlet.2012.11.058
23. Zhao Q, Bi Y, Zhong J, et al. Pristimerin suppresses colorectal cancer through inhibiting inflammatory responses and Wnt/β-catenin signaling. Toxicol Appl Pharmacol. 2020;386:114813. doi:10.1016/j.taap.2019.114813
24. Salajegheh A, Vosgha H, Rahman A, et al. Modulatory role of miR-205 in angiogenesis and progression of thyroid cancer. J Mol Endocrinol. 2015;55:183–196. doi:10.1530/JME-15-0182
25. Bi DP, Yin CH, Zhang XY, et al. MiR-183 functions as an oncogene by targeting ABCA1 in colon cancer. Oncol Rep. 2016;35:2873–2879. doi:10.3892/or.2016.4631
26. Bi WP, Xia M, Wang XJ. miR-137 suppresses proliferation, migration and invasion of colon cancer cell lines by targeting TCF4. Oncol Lett. 2018;15:8744–8748. doi:10.3892/ol.2018.8364
27. Li N, Mao D, Cao Y, et al. Downregulation of SIRT6 by miR-34c-5p is associated with poor prognosis and promotes colon cancer proliferation through inhibiting apoptosis via the JAK2/STAT3 signaling pathway. Int J Oncol. 2018;52:1515–1527.
28. Madhavan D, Peng C, Wallwiener M, et al. Circulating miRNAs with prognostic value in metastatic breast cancer and for early detection of metastasis. Carcinogenesis. 2016;37:461–470. doi:10.1093/carcin/bgw008
29. Yoshino H, Yonezawa T, Yonemori M, et al. Downregulation of microRNA-1274a induces cell apoptosis through regulation of BMPR1B in clear cell renal cell carcinoma. Oncol Rep. 2018;39:173–181. doi:10.3892/or.2017.6098
30. Wang GJ, Liu GH, Ye YW, et al. The role of microRNA-1274a in the tumorigenesis of gastric cancer: accelerating cancer cell proliferation and migration via directly targeting FOXO4. Biochem Biophys Res Commun. 2015;459:629–635. doi:10.1016/j.bbrc.2015.02.160
31. Xiang-qiang L, Shan-hong T, Zhi-yong Z, et al. [Expression and clinical significance of FOX04 in colorectal cancer]. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2011;27:969–971.
32. Liu X, Zhang Z, Sun L, et al. MicroRNA-499-5p promotes cellular invasion and tumor metastasis in colorectal cancer by targeting FOXO4 and PDCD4. Carcinogenesis. 2011;32:1798–1805. doi:10.1093/carcin/bgr213
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.