Back to Journals » OncoTargets and Therapy » Volume 10
Upregulation of long noncoding RNA LOC440040 promotes tumor progression and predicts poor prognosis in patients with prostate cancer
Authors Zhang C, Liu C, Wu J, Zheng Y, Xu H, Cheng G, Hua L
Received 30 March 2017
Accepted for publication 6 August 2017
Published 11 October 2017 Volume 2017:10 Pages 4945—4954
DOI https://doi.org/10.2147/OTT.S138354
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Geoffrey Pietersz
Cheng Zhang,1,2,* Chunlin Liu,3,* Jie Wu,2,4,* Yuxiao Zheng,2 Haoxiang Xu,2 Gong Cheng,2 Lixin Hua2
1Department of Urology, Gaoyou Traditional Chinese Medicine Hospital, Yangzhou 225600, 2Department of Urology, The First Affiliated Hospital of Nanjing Medical University, Nanjing 210029, 3Department of Urology, The Second People’s Hospital of Taizhou, Taizhou, Jiangsu, 225500, 4Department of Urology, The Affiliated Jiangning Hospital of Nanjing Medical University, Nanjing 211100, People’s Republic of China
*These authors contributed equally to this work
Background/purpose: Long noncoding RNAs (lncRNAs) play a functional role in the initiation and progression of prostate cancer (PCa). This study aimed to determine differentially expressed lncRNA through high-throughput sequencing technology and investigate its expression, biological function and clinical correlation with PCa.
Materials and methods: Candidate lncRNAs were identified through microarray screening and bioanalysis. The expression of lncRNA LOC440040 in PCa tissues and cell lines was determined by reverse-transcription polymerase chain reaction. The relationship between LOC440040 level and clinicopathological characteristics was analyzed by paired t-test or chi-square test, and its association with patient prognosis was assessed by the Kaplan–Meier method. The effects of LOC440040 on PC-3 and 22RV1 cells were evaluated by Cell Counting Kit-8 (CCK-8), migration, invasion and colony formation assays.
Results: LOC440040 expression was upregulated in PCa tissues and cell lines. Clinicopathological analysis showed that patients with high LOC440040 expression exhibited more advanced clinical features and shorter overall survival than those with low LOC440040 expression. Multivariate regression analysis revealed that LOC440040 expression was an independent prognostic factor in patients with PCa. Knockdown of LOC440040 inhibited PCa cell proliferation, migration and invasion.
Conclusion: LOC440040 may play an oncogenic role in PCa initiation and progression. This lncRNA could be a novel molecular prognostic biomarker and a potential therapeutic target for PCa.
Keywords: long noncoding RNA, LOC440040, prostate cancer, prognosis
Introduction
Prostate cancer (PCa) is the most prevalent male malignancy and the second most common cause of cancer-related deaths in men in the US.1,2 Approximately 1 million new cases of PCa are diagnosed every year, accounting for ~10% of all new tumor cases in men worldwide.3,4 PCa is usually diagnosed in men aged >65 years. This severe disease can be treated using active surveillance, prostatectomy, radiotherapy, hormonal therapy or chemotherapy.5 Thus far, the highly complex molecular mechanisms involved in PCa tumorigenesis and progression remain unclear; hence, scholars must develop novel and reliable biomarkers for early detection, effective treatment and prognosis estimation of PCa.
With the advancement of high-throughput sequencing technologies, long noncoding RNAs (lncRNAs) have become relevant to identifying human diseases, particularly cancers.6 LncRNAs are noncoding transcripts with length of >200 nucleotides and were previously considered to be transcriptional noise.7 LncRNAs have emerged as key players in several biological processes, such as cell differentiation, cell proliferation, cell apoptosis and cell cycle progression.8–13 The relationship between lncRNAs and PCa has gained increased research attention. PCa-associated ncRNA transcript 1 (PCAT-1) is a prostate-specific long noncoding RNA overexpressed in a subset of PCa and functions as a biomarker for this cancer type.14 Nuclear-enriched abundant transcript 1 (NEAT1) is an lncRNA regulated by estrogen receptor alpha and is upregulated in PCa. NEAT1 expression is associated with PCa progression and could be used as a novel biomarker of disease progression.15 Several urine-based biomarkers in PCa have been developed in preclinical studies for noninvasive detection of PCa. For example, urine PCA3 assay exhibits potential for molecular urine analysis in clinical practice. Urine PCA3 is a highly optimized, multiplex urine biomarker for accurate detection of PCa.16,17 Moreover, metastasis-associated lung adenocarcinoma transcript 1 (MALAT-1) is a promising urine biomarker for predicting PCa risks.18
Currently, the understanding of the expression and biological role of lncRNAs in PCa remains in the early phase. Thus far, the lncRNA LOC440040 in PCa has not been investigated, and its role in PCa carcinogenesis and progression remains unclear. In the present study, we examined lncRNA LOC440040 expression in PCa tissues and cell lines compared with normal tissues and cell lines. We also explored the roles of LOC440040 in the biological functions of PCa cells. Results indicated that LOC440040 may act as a potential novel molecular biomarker and therapeutic target for the treatment of PCa.
Materials and methods
Clinical specimen collection and microarray analysis
A total of 72 paired PCa tissues and carcinoma adjacent tissues were obtained from patients who underwent radical resection at The First Affiliated Hospital of Nanjing Medical University (Nanjing, Jiangsu, China) between 2010 and 2014. None of the patients had received radiotherapy, chemotherapy or endocrinotherapy prior to radical prostatectomy operation. All collected clinical tissue samples were immediately snap frozen in liquid nitrogen and stored at −80°C before RNA extraction. Three specimens were used for microarray analysis of lncRNAs, and the 69 remaining specimens were selected for additional assessments. Microarray analysis was used to identify candidate molecular targets. This study was approved by the institutional review board of the First Affiliated Hospital of Nanjing Medical University and performed in accordance with the principles of the Declaration of Helsinki. Written informed consent was obtained from each patient before operation.
Cell culture
Human PCa cell lines (PC-3, DU-145 and 22RV1) and normal myofibroblast stromal cell line (WPMY-1) were obtained from the Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences (Shanghai, China). The cells were cultured in F-12K, Roswell Park Memorial Institute (RPMI) 1640 or Dulbecco’s Modified Eagle’s Medium (DMEM; Gibco BRL, Carlsbad, CA, USA) medium supplemented with 10% heat-inactivated fetal bovine serum (FBS; Gibco BRL), 100 U/mL penicillin sodium and 100 mg/mL streptomycin sulfate at 37°C in a humidified air atmosphere with 5% CO2. Exponentially growing cells were used in all experiments.
RNA isolation, reverse transcription and quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted from PCa tissues and cell lines by using TRIzol reagent (Invitrogen Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s instructions. The isolated RNA was reverse transcribed into cDNA by using a reverse transcription kit (Takara, Dalian, China). According to the manufacturer’s protocols, reverse transcription was conducted at 37°C for 15 min, followed by 85°C for 5 s. qRT-PCR analysis was conducted using a standard protocol from Power SYBR Green (Takara). Each 10 μL of the PCR reaction volume comprised SYBR Premix (2×, 5 μL), forward primer (10 μM, 0.4 μL), reverse primer (10 μM, 0.4 μL), ROX Reference Dye II (50×, 0.2 μL), cDNA sample (1 μL) and bidistilled water (3 μL). The qRT-PCR reaction was performed on an ABI StepOnePlus instrument (Applied Biosystems, Carlsbad, CA, USA). Amplification was performed in triplicate starting with an initial denaturation step of 10 min at 95°C, followed by 50 cycles of denaturation at 95°C for 30 s, 95°C for 5 s and 61°C for 30 s and final annealing extension at 72°C for 5 min. The relative expression of lncRNA LOC440040 was calculated and normalized by 2−ΔΔCt method relative to β-actin. Each experiment was performed in triplicate. The primer sequences were as follows: 1) LOC440040: forward: 5′-ATAGTAGGGGTCATTGGGCC-3′ and reverse: 5′-AGTTGCCTTCTGTGTGTACG-3′ and 2) β-actin: forward: 5′-TCACCCACACTGTGCCCATCTACGA-3′ and reverse: 5′-CAGCGGAACCGCTCATTGCCAATGG-3′.
Small interfering RNA transfection
Three LOC440040 short interfering RNAs (siRNAs) and scrambled negative control siRNA (si-NC) were purchased from GenePharma (Shanghai, China). The target sequences for si-LOC440040 included si-LOC440040-1(5′-CCAUCAUGGAUCUGAGUGATT-3′), si-LOC440040-2(5′-CAUCUUUAUUGAGAUCUCATT-3′) and si-LOC440040-3(5′-CUGCAUUAUAAUGAAGCCATT-3′). Experimental results indicated that the latter two exhibited higher inhibition efficiency relative to the former. When the cell density in the six-well plate reached 70%, PCa cells were transfected with siRNA by using Lipofectamine 2000 (Invitrogen, Shanghai, China) according to the manufacturer’s instructions. Cells were harvested for further qRT-PCR and Western blot assays 48 h after transfection.
Cell proliferation assay
PCa cells were seeded at a density of 2,500 cells/well in 96-well plates 48 h after siRNA transfection. At 24, 48, 72 and 96 h after transfection, 10 μL of Cell Counting Kit-8 (CCK-8; Dojindo, Kumamoto, Japan) was added to the plates. The cells were incubated for 4 h at 37°C in a humidified chamber. Absorbance (optical density [OD]) was detected at a wavelength of 490 nm by using an enzyme-labeled analyzer. Each experiment was performed in triplicate.
Cell migration and invasion assay
Cell migration and invasion assays were performed using Transwell chambers (8 μm pore size; Millicell, PIEP12R48). Cells were harvested and suspended in a serum-free medium. The upper chamber was added with 200 μL of the cell suspension at a density of 3×104 cells/well. The lower chamber was added with 500 μL of the medium containing 10% FBS as a chemoattractant. After 48 h of incubation, the surface of the upper chamber was swabbed with cotton to remove cells that did not migrate. The membranes were fixed and stained with 0.5% crystal violet. Distilled water was used for washing to remove excess stain materials. Three randomly selected fields were counted per chamber by using an inverted microscope (Olympus Corporation, Tokyo, Japan). Each experiment was conducted in triplicate. The invasion assay protocol was analogous to the migration assay, except that the upper chamber was first covered with Matrigel (BD Biosciences, San Jose, CA, USA).
Colony formation assay
Cells were digested with trypsin into single-cell suspension 48 h after transfection. Briefly, 8×102 cells were seeded per well into six-well plates and maintained in the media containing 10% FBS to promote colony formation. The nutrient solution was changed every 5 days. After 14 days, the colonies were fixed with methanol for 20 min and stained with 0.1% crystal violet for 30 min. Colonies containing >50 cells were counted. Triplicate plates were measured in each group.
Western blot assay
Cells were collected 48 h after transfection and lysed using the mammalian protein extraction reagent radio immunoprecipitation assay (RIPA) (Beyotime, Beijing, China) supplemented with protease inhibitor cocktail (Roche, Shanghai, China) and phenylmethanesulfonyl fluoride (Roche). Total protein was separated by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto polyvinylidene fluoride (PVDF) membranes (Sigma-Aldrich Co., St Louis, MO, USA). The membranes were blocked with 5% nonfat milk and incubated with specific primary antihuman antibodies overnight at 4°C. The specific rabbit monoclonal antibodies for AKT, pAKT, mTOR, pmTOR, Vimentin, β-catenin and E-cadherin (1:1,000) were purchased from Cell Signaling Technology, Inc. (St Louis, MO, USA). The membranes were incubated with secondary antibodies at room temperature for 2 h. Enhanced chemiluminescent chromogenic substrate (Beyotime) was used to visualize the bands, and their intensity was quantified by densitometry (Quantity One software; Bio-Rad Laboratories Inc., Hercules, CA, USA). GAPDH was used as control.
Statistical analysis
All statistical analyses were performed using SPSS 17.0 (IBM). All data were represented as mean ± standard deviation (SD). The significance of differences among the groups was estimated by Student’s t-test or chi-square test. The Kaplan–Meier method was applied to evaluate the overall survival rates, and the log-rank test was performed to calculate differences in survival among the groups. The Cox proportional hazards regression model was used for univariate and multivariate analyses to obtain survival data. P<0.05 was considered as statistically significant.
Results
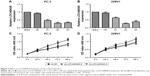
LOC440040 expression level was upregulated in PCa tissues and cell lines
In this study, a microarray containing 8,277 lncRNA probes and 32,207 mRNA probes was used to identify dysregulated lncRNA LOC440040 (Figure 1A). LOC440040 expression in 72 paired PCa tissues and adjacent nontumor tissues collected from patients with PCa were analyzed by qRT-PCR. LOC440040 expression was significantly upregulated in cancer tissues compared with that in normal tissues (P<0.05; Figure 1B). The microarray data are consistent with those of the results of qRT-PCR analysis. Furthermore, LOC440040 expression was compared between PCa cell lines (PC-3, DU-145 and 22RV1) and WPMY-1. The LOC440040 expression level significantly increased in PC-3 and 22RV1 cell lines (not shown in the DU-145 cell line, P<0.05; Figure 1C). Therefore, these two cell lines were selected for subsequent in vitro experiments.
Correlation between LOC440040 expression and clinicopathological features in patients with PCa
PCa samples were classified into high expression group (n=45) and low expression group (n=27) in accordance with the median LOC440040 expression level in all PCa samples. The association between clinicopathological features and LOC440040 expression is summarized in Table 1. The group with a high LOC440040 expression showed a higher Gleason score (P=0.027) and preoperative prostate-specific antigen (PSA) level (P=0.007) than those in the group with a low LOC440040 expression. Nevertheless, no significant correlation was found between the LOC440040 level and other clinicopathological characteristics, such as age, tumor size and clinical stage (P=0.396, 0.951 and 0.786, respectively).
  | Table 1 Correlation between LOC440040 expression and different clinicopathological features in patients with PCa |
Association between LOC440040 expression and prognosis of patients with PCa
We further investigated the associations of LOC440040 expression with survival of patients with PCa after radical prostatectomy. The Kaplan–Meier analysis indicated that patients in the high LOC440040 expression group exhibited a significantly shorter biochemical recurrence-free survival time than those in the low LOC440040 expression group (log-rank test, P<0.001; Figure 2). Univariate analysis revealed that LOC440040 expression, Gleason score, lymph node metastasis and multiple lesions were correlated with the biochemical recurrence-free survival rate of patients with PCa (P<0.05; Table 2). Multivariate analysis was performed to confirm whether these factors are independent prognostic markers for biochemical recurrence-free survival of patients with PCa (Table 2).
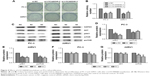
Knockdown of LOC440040 regulates PCa cell proliferation
Considering the high expression of LOC440040 in PCa tissues compared with that in adjacent nontumor tissues, we determined whether LOC440040 regulates PCa cell proliferation in vitro. Our experiments revealed that si-LOC440040-2 and si-LOC440040-3 exhibited a higher inhibitory effect on PC-3 and 22RV1 cell lines among the three si-LOC440040s (P<0.05; Figure 3A and B). CCK-8 assay was performed to determine whether knockdown of LOC440040 affects the proliferation of PC-3 and 22RV1 cells. Our data indicated that the proliferation rate of PC-3 and 22RV1 cells transfected with si-LOC440040 significantly decreased compared with that of cells transfected with si-NC and scrambled siRNA (P<0.05; Figure 3C and D).
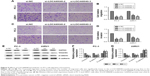
Colony formation assays were also performed to confirm the effect of LOC440040 on the proliferation of PC-3 and 22RV1 cells. Knockdown of LOC440040 suppressed the colony formation compared with that in the negative control (P<0.05; Figure 4A and B).
We further investigated whether the knockdown of LOC440040 affects the mTOR signaling pathway. The Western blots demonstrated that the protein expression levels of pAKT and pmTOR were significantly lower in PC-3 and 22RV1 cells transfected with siRNA than those transfected with si-NC. However, the variation in LOC440040 expression did not affect the expression of AKT and mTOR protein (P<0.05; Figure 4C–G). These findings suggest that LOC440040 might promote the proliferation of PCa cells in vitro and might be mediated by the PI3K/AKT/mTOR signaling pathway.
Suppression of LOC440040 inhibited PCa cell migration and invasion
We examined the effect of LOC440040 on PCa cell migration and invasion. Transwell migration assay results revealed that suppression of LOC440040 inhibited PC-3 and 22RV1 cell migration compared with that in the control group (P<0.05; Figure 5A–D). The invasive potential of si-LOC440040-treated cells was also assessed by Transwell assay using invasion chambers. The results indicated that the invasion capacity of PCa cells transfected with si-LOC440040 significantly decreased (P<0.05; Figure 4H and I). To investigate the underlying molecular mechanism of LOC440040, we analyzed the effect of LOC440040 knockdown on the epithelial mesenchymal transition (EMT) pathway. Three EMT markers, namely, Vimentin, β-catenin and E-cadherin, were selected for Western blot analysis. The relative expression levels of Vimentin and β-catenin in si-LOC440040-infected cells were lower than those in the control group. However, the expression of E-cadherin remarkably increased after LOC440040 knockdown (P<0.05; Figure 5E–G). These findings indicate that LOC440040 is a key regulator of PCa cell migration and invasion and is thus indispensable for PCa metastasis because of its role in EMT induction.
Discussion
Exploring novel molecules that participate in PCa occurrence and development is vital in improving the prevention, diagnosis and treatment of this disease. Currently, lncRNAs have been recognized to regulate complex cellular processes, such as cell growth, cell differentiation and establishment of cell identity commonly deregulated in cancer.19–21 Several lncRNAs have been identified, and their expression levels were found to be altered in PCa. For example, MALAT-1 was upregulated in human PCa tissues and cell lines and knockdown of MALAT-1 inhibited PCa features, including cell proliferation, migration, invasion and metastasis.22 Urine MALAT-1 serves as a potential novel biomarker for diagnosis of PCa, particularly for early tumor screening.18 LncRNA PCAT-1 regulates the proliferation of PCa cells through cMyc.23 SChLAP1, another lncRNA, is overexpressed in a subset of PCa. A high SChLAP1 expression by in situ hybridization (ISH) independently predicts biochemical recurrence after radical prostatectomy. SChLAP1 may be a useful tissue-based biomarker for identifying patients with PCa at a high risk of lethal progression.24 These findings indicate that lncRNA might play a fundamental role in PCa initiation and development and thus exhibit potential for clinical application.
Advances in high-throughput technology have enabled the identification of unbiased biomarkers and therapeutic targets.25 LOC440040 is a novel identified lncRNA located in the chromosome 11p11.12 gene desert. Microarray data analysis reveals that LOC440040 is overexpressed in PCa tissues. In the present study, we first analyzed LOC440040 expression in PCa tissues and cell lines through qRT-PCR. Consistent with the microarray data, LOC440040 expression significantly increased in PCa specimens compared with that in adjacent nontumor tissues. Furthermore, LOC440040 expression was upregulated in PCa cell lines (PC-3, 22RV1) compared with that in WPMY-1 cell. These findings provide the first evidence that LOC440040 possibly plays an oncogenic role in PCa carcinogenesis. We then explored whether LOC440040 expression is significantly associated with clinicopathological characteristics of patients with PCa. LOC440040 was highly expressed in patients with high Gleason score and preoperative PSA level. Additionally, our results indicate that patients with a high LOC440040 expression tend to have lower biochemical recurrence-free survival rate than those with a low expression. Finally, multivariate Cox regression analysis indicated that a high LOC440040 expression could be an independent indicator of unfavorable prognosis for patients with PCa. To the best of our knowledge, this study is the first to investigate LOC440040 expression and its clinical significance in PCa.
In vitro experiments were performed to elucidate the biological functions of LOC440040 in PCa. We detected cell proliferation, colony formation, migration and invasion in PCa cell lines. Our results demonstrate that siRNA-mediated knockdown of LOC440040 in PC-3 and 22RV1 cells inhibited cell proliferation, colony formation, migration and invasion in PCa. In comparison with other lncRNAs reported in PCa, LOC440040 exhibits the following distinct features: 1) LOC440040 expression is upregulated in PCa tissues and cell lines; 2) a high LOC440040 expression is associated with a low biochemical recurrence-free survival rate and 3) the proliferation and metastasis of PCa cells are inhibited after knockdown of LOC440040. Hence, lncRNA LOC440040 could act as an oncogene in the development of PCa.
The PI3K–AKT–mTOR signaling pathway regulates cellular functions, including cell growth, proliferation, metabolism, survival and angiogenesis.26 To determine the molecular mechanism through which LOC440040 controls cell proliferation, we determined the effect of LOC440040 knockdown on PI3K/Akt/mTOR. The results suggested that the PI3K/Akt/mTOR signaling pathway remarkably decreased relative to the inner protein expression level after LOC440040 knockdown. Hence, LOC440040 possibly contributes to PCa tumorigenesis by regulating the PI3K/Akt/mTOR signaling pathway. Multiple lines of evidence show that EMT-like events are important for tumor progression and malignant transformation. These events confer incipient cancer cells with invasive and metastatic properties.27,28 In the present study, we explored the effect of LOC440040 knockdown on EMT in PCa. Our data illustrated that the upregulation of LOC440040 promoted the migration and invasion of PCa cells in vitro. After knockdown of LOC440040, the protein expression levels of Vimentin and β-catenin were downregulated. Thus, we may deduce that LOC440040 is necessary for PCa metastasis possibly because it induces EMT.
However, this research still presents some limitations. First, clinical data were collected using a retrospective approach. As such, future studies should use a large sample size to validate our results. Second, the relationship between LOC440040 and cell cycle and xenograft carcinogenesis was not investigated, despite that we analyzed the effects of LOC440040 on the proliferation, colony formation, migration and invasion of PCa cells. Third, according to the results of microarray data, THBS2 and INHBA might be the target genes of LOC440040. Therefore, comprehensive studies must be performed to elucidate the precise mechanisms through which the genesis and development of PCa are promoted.
Collectively, this study is the first to explore the function of LOC440040 in PCa progression. The results demonstrate that intervention of LOC440040 function by drugs exhibits potential clinical value in the prevention of PCa. Therefore, our findings may not only enrich our understanding regarding the basic molecular mechanism underlying the role of LOC440040 in PCa but also provide a potential novel therapeutic target and promising prognostic indicator for future treatment of PCa.
Conclusion
This study suggested that LOC440040 expression is increased in PCa tissues and cell lines. Our results also indicated that a high LOC440040 expression level was associated with tumor progression and poor survival rate in PCa patients. The effects of LOC440040 on cell proliferation, cell migration and invasion showed that it contributes to tumorigenesis in PCa. A high LOC440040 expression promotes PCa cell growth, invasion and metastasis via the induction of PI3K/Akt/mTOR signaling pathway and EMT process. These findings indicated that LOC440040 may exert carcinogenic functions in the genesis and progression of PCa, providing a potential attractive therapeutic target and prognostic factor for this malignancy.
Acknowledgment
This study was supported by the Natural Science Foundation of Jiangsu Province (BK 20141495).
Disclosure
The authors report no conflicts of interest in this work.
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. | ||
Fu W, Yao J, Huang Y, et al. LXR agonist regulates the carcinogenesis of PCa via the SOCS3 pathway. Cell Physiol Biochem. 2014;33(1):195–204. | ||
Chang ET, Boffetta P, Adami HO, Cole P, Mandel JS. A critical review of the epidemiology of Agent Orange/TCDD and prostate cancer. Eur J Epidemiol. 2014;29(10):667–723. | ||
Luo G, Wang M, Wu XC, et al. Long non-coding RNA MEG3 inhibits cell proliferation and induces apoptosis in prostate cancer. Cell Physiol Biochem. 2015;37(6):2209–2220. | ||
Westdorp H, Skold AE, Snijer BA, et al. Immunotherapy for prostate cancer: lessons from responses to tumor-associated antigens. Front Immunol. 2014;5:191. | ||
Fatima R, Akhade VS, Pal D, Rao SM. Long noncoding RNAs in development and cancer: potential biomarkers and therapeutic targets. Mol Cell Ther. 2015;3:5. | ||
Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1(5):391–407. | ||
Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–1076. | ||
Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9(6):703–719. | ||
Guttman M, Donaghey J, Carey BW, et al. LincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477(7364):295–300. | ||
Wang Y, Gao S, Liu G, Jia R, Fan D, Feng X. Microarray expression profile analysis of long non-coding RNAs in human gastric cardiac adenocarcinoma. Cell Physiol Biochem. 2014;33(4):1225–1238. | ||
Liu H, Song G, Zhou L, et al. Compared analysis of LncRNA expression profiling in pdk1 gene knockout mice at two time points. Cell Physiol Biochem. 2013;32(5):1497–1508. | ||
Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10(3):155–159. | ||
Prensner JR, Iyer MK, Balbin OA, et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol. 2011;29(8):742–749. | ||
Chakravarty D, Sboner A, Nair SS, et al. The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat Commun. 2014;5:5383. | ||
Laxman B, Morris DS, Yu JJ, et al. A first-generation multiplex biomarker analysis of urine for the early detection of prostate cancer. Cancer Res. 2008;68(3):645–649. | ||
Hessels D, Klein-Gunnewiek JM, Van OI, et al. DD3(PCA3)-based molecular urine analysis for the diagnosis of prostate cancer. Eur Urol. 2003;44(1):8–15. | ||
Wang FB, Ren SC, Chen R, et al. Development and prospective multicenter evaluation of the long noncoding RNA MALAT-1 as a diagnostic urinary biomarker for prostate cancer. Oncotarget. 2014;5(22):11091–11102. | ||
Hu W, Alvarez-Dominguez JR, Lodish HF. Regulation of mammalian cell differentiation by long non-coding RNAs. EMBO Rep. 2012;13(11):971–983. | ||
Flynn RA, Chang HY. Long noncoding RNAs in cell-fate programming and reprogramming. Cell Stem Cell. 2014;14(6):752–761. | ||
Rossi MN, Antonangeli F. LncRNAs: new players in apoptosis control. Int J Cell Biol. 2014;2014:473857. | ||
Ren SC, Liu YW, Xu WD, et al. Long noncoding RNA MALAT-1 is a new potential therapeutic target for castration resistant prostate cancer. J Urol. 2013;190(6):2278–2287. | ||
Prensner JR, Chen W, Han SM, et al. The long non-coding RNA PCAT-1 promotes prostate cancer cell proliferation through cMyc. Neoplasia. 2014;16(11):900–908. | ||
Mehra R, Udager AM, Ahearn TU, et al. Overexpression of the long non-coding RNA SChLAP1 independently predicts lethal prostate cancer. Eur Urol. 2015;70(4):549–552. | ||
Zhou D, Teng F, Verhaak RG, et al. Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer. Nat Struct Mol Biol. 2013;20(7):908–913. | ||
Riquelme I, Tapia O, Espinoza JA, et al. The gene expression status of the PI3K/AKT/mTOR pathway in gastric cancer tissues and cell lines. Pathol Oncol Res. 2016;22(4):797–805. | ||
Makker A, Goel MM. Tumor progression, metastasis, and modulators of epithelial-mesenchymal transition in endometrioid endometrial carcinoma: an update. Endocr Relat Cancer. 2016;23(2):R85–R111. | ||
Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2(6):442–454. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.