Back to Journals » Cancer Management and Research » Volume 10
Upregulation of KIF20A correlates with poor prognosis in gastric cancer
Authors Sheng Y, Wang W , Hong B , Jiang X , Sun R, Yan Q, Zhang S, Lu M , Wang S, Zhang Z, Lin W, Li Y
Received 2 June 2018
Accepted for publication 2 August 2018
Published 23 November 2018 Volume 2018:10 Pages 6205—6216
DOI https://doi.org/10.2147/CMAR.S176147
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Antonella D'Anneo
Yi Sheng,1,* Wei Wang,2,* Bo Hong,2 Xingwang Jiang,1 Ruochuan Sun,1 Qiang Yan,1 Shangxin Zhang,1 Mingdian Lu,1 Shengyi Wang,1 Zhen Zhang,1 Wenchu Lin,2 Yongxiang Li1
1Department of General Surgery, The First Affiliated Hospital of Anhui Medical University, Hefei, Anhui, China; 2High Magnetic Field Laboratory, Chinese Academy of Sciences, Hefei, Anhui, China
*These authors contributed equally to this work
Background: KIF20A is well known as one of the key proteins in mitosis. Recently, a number of studies illustrated that KIF20A might function as an oncogene in some carcinomas. However, its expression levels and clinical value remained unclear in gastric cancer (GC).
Patients and methods: In this study, we investigated the expression of KIF20A in samples from GC patients and cell lines by quantitative real-time PCR and Western blot. The function of KIF20A in cell proliferation of GC cell lines was examined via cell viability and colony formation assays. Immunohistochemistry assay based on a tissue microarray consisting of 146 cases was performed to evaluate the prognostic value of KIF20A. The overall survival rate of 122 GC patients based on KIF20A expression was analyzed as well. Finally, using KIF20A inhibitor, genistein, and combining it with cisplatin or fluorouracil, the antitumor effects were studied.
Results: Most GC samples (56.76%) showed higher KIF20A expression level compared to their corresponding normal specimens, which demonstrated the potential oncogenic role of KIF20A in GC. The functional studies elucidated the essential role of KIF20A in GC cell proliferation. Besides, tissue microarray result showed that the expression level of KIF20A was significantly related to the histological grades (P=0.036). Furthermore, we found the expression of KIF20A was related to poor overall survival rate, which is coincident with the results from Kaplan–Meier plotter database. In addition, we found that a KIF20A inhibitor, genistein, could enhance the antitumor activity of cisplatin and fluorouracil, which might be considered as a chemosensitive agent in GC.
Conclusion: KIF20A can promote cell proliferation in GC, which might be used as an independent prognostic factor and a potential therapeutic target.
Keywords: KIF20A, genistein, gastric cancer, prognosis
Introduction
Gastric cancer (GC) is one of the frequently diagnosed cancers worldwide.1 In China, GC is the second most commonly diagnosed cancer and the second leading cause of cancer death.2 Despite the declining rates in recent years, which benefited from the changes in diet and improved medical care, the prognosis of advanced GC patients remains poor. Therefore, the development of new therapeutic targets is still urgently required.
Kinesin superfamily (KIF) proteins, which possess an ATPase activity and motion characteristics in most members,3 participate in numerous cellular biological activities, including mitosis, meiosis, and the transport of macromolecules.4 A total of 45 KIF proteins with varying functions have been identified and classified into 14 subfamilies based on structural differences.5,6 KIF20A, a member of kinesin-6 family, was identified as a motor protein required for coupling actin and microtubule cytoskeletons that ensure the spatial coordination between the forming of Golgi/trans-Golgi network membranes by RAB6-positive vesicles fission and their exit along microtubules.7 Furthermore, KIF20A has a microtubule-dependent plus-end motility and, therefore, has been involved in different cellular processes such as formation of the mitotic spindle and chromosome partitioning.8
Besides its important functions in mitosis, KIF20A was first found to be overexpressed in pancreatic ductal adenocarcinoma (PDAC).9 When downregulated the endogenous KIF20A in PDAC cell lines by small interfering RNA, the cell growth was drastically inhibited.9 This result indicated that KIF20A might work as an oncogene in PDAC. Further investigation in human hepatoma cell lines exhibited high KIF20A levels as well, whereas it was undetectable in normal human hepatocytes.10 RNAi-mediated KIF20A knockdown in hepatoma cells induced polyploidization, which is consistent with its essential function in promoting cytokinesis and inhibition of cell proliferation without inducing apoptosis.10 KIF20A was first noticed in GC because of a significant decrease in its expression by vorinostat treatment, which induced the apoptosis and autophagy of GC cells.11 Furthermore, Yan et al12 showed that the cell viability of GC SGC7901 cells was inhibited by KIF20A RNAi, which illustrated the important role of KIF20A. Afterward, peptide derived from KIF20A was used as immunotherapy vaccine alone or combined with other peptides/chemotherapy drugs to treat GC, which achieved remarkably higher overall survival (OS) rate.13–16 These studies revealed that KIF20A should be a promising therapeutic target for cancers. However, the expression of KIF20A and its role as well as the clinical values in GC still remain unclear.
Genistein, one of the most important isoflavones, has been shown to have anticancer effects in many cancers, without exerting toxic effects on normal cells.12,17–20 In GC, Yan et al12 showed that genistein can induce mitotic arrest by downregulation of KIF20A, which inhibited cell viability and induced G2/M arrest. Therefore, genistein might be a potential drug targeting KIF20A in GC. In our study, we tested its antitumor effects in GC cell lines.
Notably, an increasing number of studies have revealed that genistein inhibits the proliferation of cancer cells by cooperatively or synergistically enhancing the effect of antitumor drugs. It has been reported that genistein promotes the antitumor effect in coordination with tamoxifen in estrogen receptor-positive breast cancer cells,21 with 5-fluorouracil in colon cancer cells,22 and with cisplatin in breast and colon cancer cells.23 According to the National Comprehensive Cancer Network clinical practice guidelines in oncology (2017), fluorouracil and cisplatin are the first-line therapy in GC. Therefore, we examined if genistein could sensitize GC cells to fluorouracil or cisplatin treatment.
In this study, we initially determined high KIF20A expression status in human GC tissues and cell lines. We found that high levels of KIF20A could promote cell proliferation. Importantly, this high expression level was significantly correlated with clinicopathologic characteristics and poor prognosis. Genistein can inhibit the cell growth and enhance the antitumor effect of fluorouracil or cisplatin against KIF20A high-expression GCs.
Patients and methods
Materials
Genistein, cisplatin, and fluorouracil were purchased from Selleck Chemical (Shanghai, China), and stock solutions were prepared in dimethyl sulfoxide (Sigma-Aldrich, St Louis, MO, USA) at a concentration of 50, 10, and 10 mM, respectively. Antibody for KIF20A was purchased from Santa Cruz Biotechnology (Dallas, TX, USA). Actin antibody was obtained from TransBionovo (Beijing, China).
Patients and tissue samples
After obtaining written informed consent from all the patients and approval of the Ethics Committee of Anhui Medical University, tissue specimens were collected and immediately preserved in liquid nitrogen in the First Affiliated Hospital of Anhui Medical University (Hefei, China). One hundred and twenty-two GCs and 24 normal gastric mucosa tissues were subjected to immunohistochemistry (IHC). Thirty-seven GCs and their corresponding adjacent normal tissues were examined by quantitative real-time PCR (qRT-PCR), in which 12 pairs were selected and detected by Western blot. None of the patients received preoperative treatment. According to the TNM staging guide (2010) released by the American Joint Committee on Cancer, pathological diagnosis was identified by at least two pathologists.
Cell culture and lentivirus infection
This study was approved by the Ethics Committee of Anhui Medical University. Five human GC cell lines (AGS, SGC-7901, BGC-823, NCI-N87, and MGC-803) and immortalized human gastric mucosal epithelial cell line GES-1 were maintained in RPMI-1640 medium supplemented with 10% FBS and 1% penicillin/streptomycin in a humidified incubator at 37°C with 5% CO2. AGS cell line was derived from the Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences. The other five cell lines were kindly supplied by Dr Chunwei Peng from the Department of General Surgery, the First Affiliated Hospital of Anhui Medical University. RPMI-1640 medium, FBS, and penicillin/streptomycin were purchased from Thermo Fisher Scientific (Waltham, MA, USA).
KIF20A overexpression lentiviral vector GV358 (KIF20A-OE, Ubi-KIF20A-3FLAG-SV40-EGFP-IRES-puromycin) and GV358 empty vector (CON238, Ubi-MCS-3FLAG-SV40-EGFP-IRES-puromycin) were obtained from Genechem (Shanghai, China). GV248 vector was used to interfere with KIF20A expression; the shRNA target sequences for human KIF20A (hU6-KIF20A-Ubiquitin-EGFP-IRES-puromycin) were as follows: shKIF20A-1: 5′-CAGAAGAATATAAGGCTGT-3′; shKIF20A-2: 5′-GTTCCTGCATGATTGTCAA-3′; shKIF20A-3: 5′-GATGACGATGTCGTAGTTT-3′. CON077 (hU6-MCS-Ubiquitin-EGFP-IRES-puromycin) with the target sequence of 5′-TTCTCCGAACGTGTCACGT-3′ served as a control. The procedures of lentivirus packaging and infection were performed as described previously.24 Overexpression and knockdown of target genes efficacy were analyzed by Western blot.
RNA extraction and qRT-PCR analysis
Total RNA was extracted from the 37 pairs of fresh specimens or cells using TransZol Up Plus RNA Kit (Transgen, Beijing, China). After synthesizing the first strand cDNA via RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific), qRT-PCR was performed in triplicate using FastStart Essential DNA Green Master reactions (Roche, Mannheim, Germany) on a Roche LightCycler 96 Real-Time PCR System (Roche). The primer sequence for KIF20A detection was as follows: forward primer 5′-ACTTTGCGGCTATGCGAGGAT-3′ and reverse primer 5′-TGAGAAGATGCTGTGACTGCG-3′. The reaction conditions were as follows: denaturation at 95°C for 10 minutes, followed by 40 cycles of 95°C for 15 seconds, 60°C for 20 seconds, and 72°C for 20 seconds, with a final extension at 72°C for 30 seconds. β-Actin served as the internal control with the primers: forward 5′-TCTGGCACCACACCTTCTACA-3′ and reverse 5′-AGGGATAGCACAGCCTGGATA-3′. The relative expression of each sample was calculated using the 2−ΔΔCT method.
Western blot
By homogenization in lysis buffer (150 mM NaCl, 50 mM Tris-HCl pH 8.0, 1% Triton X-100, and 1 mM EDTA), proteins were extracted from tissues or cells and quantified using BCA Protein Assay Kit (Beyotime, Shanghai, China). Then the proteins from each sample were separated by SDS-PAGE and transferred onto Immobilon P membrane (EMD Millipore, Billerica, MA, USA). After hybridization with a primary antibody followed by secondary antibody incubation, the signals on the membranes were visualized using SuperSignal West Pico Chemiluminescent substrate (Thermo Fisher Scientific).
Cell viability assay
One thousand cells infected by different lentiviruses or treated with different drugs were seeded into 96-well plates and cultured for 72 hours. Then the cell viability was examined via CellTiter-Glo Luminescent assay (Promega, Madison, WI, USA) according to the manufacturer’s instructions. Luminescence was measured in a multi-label plate reader (Envision; PerkinElmer, Waltham, MA, USA). Data were represented by the mean of three independent measurements with standard error of mean (SEM). The IC50 of genistein was calculated using the nonlinear regression inhibitor vs response curve from GraphPad Prism 6 software (GraphPad Software, Inc., La Jolla, CA, USA).
Colony formation assays
A total of 3,000 cells infected by different lentiviruses were seeded into six-well plates. After 7 days of incubation, colonies were fixed with 4% paraformaldehyde for 15 minutes at room temperature and then washed with PBS. Then 0.5% crystal violet was used to stain the fixed cells for 30 minutes, followed by the washing step under tap water. Then the plates were left to dry overnight. Finally, the number of colonies (>50 cells/colony) was counted. Each assay was repeated at least three times.
Immunohistochemistry
Before IHC, H&E-stained slides were screened to identify optimal intratumoral tissue for analysis. Multiple 4 μm sections were cut with a Micron microtome, and then the sections were baked at 63°C for 1 hour, deparaffinized with xylene, and rehydrated through graded ethanol solutions and finally washing in distilled water. Antigen retrieval was performed by placing the sections in 0.01 M citrate buffer pH 6.0 in a pressure cooker for 5 minutes at 120°C. After antigen retrieval, the endogenous peroxidase activity of the sections was quenched by 3% hydrogen peroxidase (H2O2) in methanol. To inhibit nonspecific antigen–antibody reactivity in the IHC assay, the sections were blocked by 1% BSA. Then the slides were incubated with the primary antibody, anti-KIF20A antibody (1:2,000; Thermo Fisher Scientific) overnight at 4°C followed by washing. Thereafter, the sections with bound primary antibody were detected by horseradish peroxidase-labeled anti-rabbit IgG secondary antibodies at room temperature for 30 minutes, followed by washing and visualization using an autostainer link instrument, and then staining was performed. The IHC stained slides were scored by two independent experienced pathologists who had no prior knowledge of the clinicopathologic information of these patients. We classified immunoreactivity in cells based on staining as follows: negative, nucleus only stained, cytoplasm only stained, nucleus and cytoplasm both stained. We categorized these subgroups as nucleus negative and positive, and cytoplasm negative and positive. According to the dominant staining intensity of GC mucosa cells and normal mucosa cells, the scores were classified into four grades based on the percentage of staining (0 points for no cells stained, 1 point for <25% of cells stained, 2 points for 25%–75% of cells stained, and 3 points for >75% of cells stained), and the staining intensity of immunoreactivity was graded on a scale of 0–3 (intensity scores): negative (0), weak (1), moderate (2), and strong (3). The immunoreactivity score (IRS) was defined as multiplication of both parameters. Specimens were scored as follows: negative or weak (−, IRS=0–2), positive (+, IRS=3–9).
Data mining
Data mining and analysis of KIF20A mRNA expression were conducted using data sets from the Oncomine.25 The prognostic value of KIF20A mRNA expression was retrieved from Kaplan–Meier Plotter.26
Statistical analysis
GraphPad Prism 6 software (GraphPad Software, Inc.) was used to present graphs. Statistical analyses were conducted using SPSS 22.0 software (IBM Corporation, Armonk, NY, USA). The differential expression of mRNA between paired fresh frozen GCs and normal tissues was analyzed by paired t-test. The relations between KIF20A expression and clinicopathologic characteristics were assessed by the Pearson chi-squared test. OS was analyzed by Kaplan–Meier method, and the significant differences of survival curves were displayed by the log-rank test. Multivariate survival analysis was performed using the Cox proportional hazards regression model. All statistical tests were two sided. P<0.05 was considered statistically significant. *, **, ***, and **** corresponded to P<0.05, P<0.01, P<0.001, and P<0.0001, respectively.
Results
KIF20A is upregulated in GC
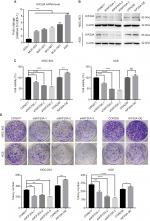
In order to investigate the expression level of KIF20A in GC, we first performed qRT-PCR in a cohort of 37 fresh GC tissues together with their corresponding adjacent normal tissues. The mRNA expression level of KIF20A was significantly higher in 56.76% (21/37) of GC tissues when compared with their matched controls (Figure 1A). Subsequently, among the KIF20A upregulated samples, 12 were selected and their protein expression levels were examined. They were upregulated in GC tissues at the translation level as well (Figure 1B). Furthermore, in the Oncomine database, the mRNA expression profiles of KIF20A in GC and normal tissues were analyzed, whose results were consistent with our study (Figure 1C). In addition, KIF20A was overexpressed in GC MGC-803, BGC-823, NCI-N87, SGC-7901, and AGS cell lines when compared with immortalized human gastric mucosal epithelial cell line GES-1 (Figure 2A). Taken together, our results demonstrate that KIF20A is upregulated in GC at both mRNA and protein levels.
  | Figure 1 mRNA and protein expression levels of KIF20A in gastric cancer tissues and the corresponding normal tissues. Notes: (A) Relative mRNA expression of KIF20A was detected by quantitative real-time PCR and is shown in box and whisker plot. (B) Protein expression of KIF20A was estimated by Western blot assay. 1–12 correspond to 12 samples from GC patients. (C) Three mRNA expression profiles of KIF20A in gastric carcinoma and normal tissues27–29 were extracted from Oncomine and are presented as a box and whisker plot. Abbreviations: N, normal control; T, tumor tissue. |
KIF20A affects cell proliferation in GC cells
To explore the effect of KIF20A on the proliferation of GC cells, we adopted shRNA constructs to repress KIF20A as well as overexpression construct to increase the expression of KIF20A and examined their functions. AGS and MGC-803 cell lines, in which KIF20A was the most and the slightest upregulated comparing to normal control (Figure 2A), were chosen for the functional studies. The knockdown or overexpression efficiency was first validated after 48 hours of infection by lentivirus. Western blot assay showed that the protein levels of KIF20A were reduced by all three shRNAs in AGS and MGC-803, respectively (Figure 2B). Compared with the empty vector CON077, shKIF20A-2 and shKIF20A-3 significantly decreased the KIF20A protein level in both cell lines. When transfected with the overexpression vector, slight increase in KIF20A was identified in both cells (Figure 2B), which might be due to the inhibitory effect of their high endogenous protein level. We thereby examined whether KIF20A expression affected cells’ short-term viability (72 hours) and long-term proliferative potential (7 days). As shown in Figure 2C, knockdown of KIF20A via shRNAs dramatically inhibited cell viability in both cell lines. Meanwhile, KIF20A overexpression slightly promoted cell viability in MGC-803, but no significant difference was found in AGS. Considering AGS has the highest KIF20A expression level, the additional overexpression may show little effect. Furthermore, the long-term proliferative potential shown by KIF20A was observed by colony formation assay. Consistent with the cell viability analysis, knockdown of KIF20A inhibited the colony formation, whereas KIF20A overexpression group showed more colonies (Figure 2D). These results indicate that KIF20A expression is positively associated with the growth of GC cells.
Upregulation of KIF20A is significantly associated with the clinical prognosis of GC patients
As high KIF20A level promotes GC cell proliferation, it may lead to the rapid progression of tumors and influence the prognosis of patients. Therefore, we further investigated whether GC patients with overexpressed KIF20A had a poorer prognosis than KIF20A-negative patients. A tissue microarray (TMA) consisting of 122 GC tissues and 24 normal gastric tissues was conducted. As shown in Figure 3A, B, strong staining of KIF20A was observed in 54.92% (67/122) of GC tissues and in 29.17% (7/24) of normal gastric tissues (P<0.05). Within the tumor, KIF20A staining was predominantly found in the cytoplasm. Less than 10% of the cancer cells also showed nuclear KIF20A positivity. In 45% of the GC samples, KIF20A staining intensity was weak to moderate. Chi-squared test was conducted to detect the associations between the protein expression level of KIF20A and several clinicopathologic variables of GC patients (Table 1). Analysis showed that the protein expression level of KIF20A was significantly related to the histological grades (P=0.036). While compared with the well-differentiated (low-grade) and moderately differentiated (intermediate-grade) samples, KIF20A protein expression was significantly higher in poorly differentiated and anaplastic samples that belonged to malignant grades. However, the expression of KIF20A had no significant relationship with gender, age, tumor diameter, depth of invasion, tumor location, lymph node metastasis, and TNM stage (Table 1).
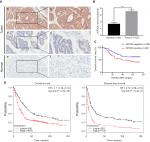  | Figure 3 Survival analysis of patients with GC based on KIF20A expression. Notes: (A) Representative images of KIF20A in GC tissues and adjacent normal tissues: (a, b) high KIF20A level in cancerous epithelia; (c, d) low KIF20A level in cancerous epithelia; (e, f) low KIF20A level in normal gastric epithelia. Magnification: (a, c, e) 100× and (b, d, f) 200×. (B) Comparison of KIF20A immunoreactivity of GC tissues (n=122) and normal gastric tissues (n=24) in the TMA. The immunoreactivity is presented as mean±SEM. ***P < 0.001. (C) The overall survival analysis based on KIF20A expression in the TMA. (D) The prognostic value of KIF20A level in GC patients from Kaplan–Meier plotter database.26 Abbreviations: GC, gastric cancer; SEM, standard error of the mean; TMA, tissue microarray. |
  | Table 1 Relationships between KIF20A protein expression and various clinicopathologic variables in GC patients Note: Bold value denotes P-value < 0.05. Abbreviation: GC, gastric cancer. |
Next, we assessed the clinical prognosis based on log-rank test in survival analysis. As shown in Table 2, we found the depth of invasion, histological grade, lymph node metastasis, TNM stage, and KIF20A expression were statistically significant factors for OS in univariate analysis. Then, the multivariate Cox regression analysis was performed on these factors, except for depth of invasion and lymph node, which were included in TNM stage (Table 2). The results further supported them to be independent prognostic factors. Median survival time for patients with higher KIF20A expression (40.16±24.65 months) was much shorter than that of patients with lower KIF20A expression (48.91±21.84 months; Figure 3C). Further examination of the results from a larger sample size (593 and 359 samples for OS and disease-free survival assay, respectively) using Kaplan–Meier plotter indicated that the GC patients with a higher mRNA level of KIF20A had much worse OS and disease-free survival rate (Figure 3D). Generally, these results indicated that KIF20A could be an independent prognostic factor for GC.
Genistein combined with cisplatin or fluorouracil can be candidate therapies for GC
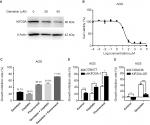
Our above results suggest that KIF20A could promote GC proliferation and is related to poor prognosis. KIF20A may represent a molecular target for the treatment of GC. A previous study suggested that KIF20A was significantly downregulated by genistein in a dose-dependent manner.12 So, we first examined the killing effect of genistein in AGS, the highest KIF20A level among the five GC cell lines. As expected, the expression of KIF20A was inhibited by genistein in a dose-dependent manner (Figure 4A). The concentration of genistein that had cell growth inhibition effect was in a micromolar range as its IC50 value was about 20 µM (Figure 4B). Although numerous reports verified that there were no toxic effects exerted by genistein to normal cells,12,17–20 its dose for inhibition tumor cell growth (nearly 100 µmol range) was relatively high and had potential risk in clinical application. So, we thought whether combining it with the first-line chemotherapy drugs in GC could enhance their antitumor effects. Therefore, we tested the cell growth inhibition effects of genistein together with cisplatin or fluorouracil. As shown in Figure 4C, AGS cells were treated with dimethyl sulfoxide control, genistein (10 µM), cisplatin (4 µM), fluorouracil (0.5 µM), genistein/cisplatin combination, and genistein/fluorouracil combination for 72 hours. Cell viability assay showed that cotreatment of genistein and cisplatin remarkably inhibited cell growth when compared with the single agents alone, indicating additive inhibitory effect in AGS cells (Figure 4C). Meanwhile, genistein also could enhance the antitumor activity of fluorouracil in AGS cells (Figure 4C). Then, we further examined the killing effect of cisplatin and fluorouracil in shKIF20A-1 AGS cells and obtained almost the same results as genistein combination experiments (Figure 4D). Overexpression KIF20A could rescue the cell growth inhibition by genistein (Figure 4E). These results demonstrated that genistein could inhibit the expression of KIF20A which repressed the growth of GC tumor cells, and downregulation of KIF20A could enhance the antitumor effect of first-line chemotherapy drugs cisplatin and fluorouracil in GC. Taken together, our study suggests that combination of KIF20A inhibitors such as genistein with cisplatin or fluorouracil might be an effective regimen for GC treatment.
Discussion
The survival of eukaryotes depends on the accurate coordination of mitosis with cytokinesis. Previous studies indicated that KIF20A regulates mitosis and cytokinesis.30,31 For instance, KIF20A is essential for recruitment of PLK1 and chromosomal passenger complex to the central spindle in anaphase, which are key regulators of chromosome segregation and cytokinesis.32–34 It suggests that dysregulated KIF20A may cause abnormal cell division and the generation of binucleated cells, leading to chromosomal aneuploidy and genomic instability in cancer.35–37 Recent research has implied that overexpression of KIF20A is associated with a number of cancers including GC.9–11,38 However, the expression pattern and the function of KIF20A, as well as its association with the clinicopathologic features in GC are still unclear.
Therefore, we first explored the expression level of KIF20A in 37 GC patients and found that KIF20A expression is generally upregulated at both transcriptional and translational levels compared to its corresponding normal tissues. We noticed that some normal tissues had higher KIF20A expression as well. This might be due to the cutting positions of the normal tissues were too close to the tumors and some KIF20A-positive cells were mixed inside or its indeed upregulated in the normal gastric mucosa tissues in these patients. Thus, it is really interesting to study if these highly KIF20A basal expression level patients have different clinical features, especially poor prognosis, which we will illustrate in the future studies.
To investigate whether its overexpression is correlated with GC cell proliferation, we performed cell viability and colony assay using lentivirus-mediated up- and downregulation of KIF20A in GC cell lines. Notably, knockdown of KIF20A can significantly suppress cell proliferation, whereas overexpression of KIF20A can increase cell proliferation. AGS has the highest KIF20A expression level; in contrast, the lowest expression cell line is MGC-803 among the five GC cell lines selected by us. Unlike the effective knockdown, the overexpression was not significant, which might be due to the transfection efficiency of lentivirus or the inhibition effect of high endogenous KIF20A protein level. Due to the lower endogenous level of KIF20A in MGC-803, the overexpression was a little higher than in AGS, which led to more cell proliferation and colony formation. Although the silencing of KIF20A inhibited cell viability through induction of G2/M arrest,12 the functional factors and pathways still remain unknown. Especially, the molecular mechanisms for the cell proliferation enhanced by KIF20A overexpression are worth investigating. Therefore, KIF20A plays a key role in GC cell proliferation.
These above findings prompted us to question the role of KIF20A in clinical features of GC. Analysis of TMA identified that GC patients with positive KIF20A expression showed higher differentiation in malignant grades; in contrast, no relationship was found with other clinicopathologic features such as depth of invasion, lymph node metastasis, and TNM stages. These results showed that KIF20A expression might only lead to the proliferation of tumor and has no relationship to tumor invasion and metastasis. Based on this hypothesis, KIF20A might be a valuable therapeutic target for GC proliferation. Kaplan–Meier analysis based on the TMA and database revealed that patients with positive KIF20A expression had a shorter OS. These results indicate that KIF20A could serve as a useful prognostic biomarker for GC. Interestingly, the OS rate in our patients was longer than in GC patients from the database. This difference might be due to the races of the patients. Our samples were from Chinese population and the patients studied in the database were almost from Western countries. However, whether Chinese GC patients have longer OS rate needs to be studied in detail.
Cisplatin and fluorouracil are the first-line chemotherapy drugs used clinically for GC. However, the strong toxicity and side effects limit their application. So, researchers want to find new agents which could make the GC sensitized to these chemotherapy drugs. Various reports proved that genistein, a KIF20A inhibitor, is safe for human health and it is considered as a promising chemopreventive agent to various types of cancer.39 Genistein has a potential chemosensitive role, which shows synergistic effect with a number of widely used antitumor drugs such as docetaxel, tamoxifen, 5-fluorouracil, and cisplatin.21–23,40,41 However, its role in GC had not been studied much. Therefore, we combined genistein with cisplatin or fluorouracil to treat the GC cells. The results showed that genistein had additive effect when treated together with cisplatin or fluorouracil in GC. To summarize, genistein could be considered as a promising enhancing agent of cisplatin and fluorouracil for the treatment of GC.
Conclusion
Using specimens from patients, our study has shown that KIF20A is upregulated at both mRNA and protein levels in GC. Dysregulation of KIF20A significantly affects proliferation of GC cell lines, which demonstrates its important role in GC. Besides, the prognostic significance of KIF20A expression assays revealed that it is significantly associated with worse histological grades and acts as an independent prognostic factor for the OS of patients with GC. In addition, we illustrated that genistein, a drug inhibiting KIF20A expression, showed an enhanced anticancer effect with cisplatin or fluorouracil in GC, which might be helpful to develop new strategies to conquer GC. Taken together, we suggest that KIF20A should be considered as a candidate prognostic factor and therapeutic target for GC.
Acknowledgments
We are grateful to all patients enrolled in this study. We thank the members of the Lin laboratory for critical reading of the manuscript and helpful discussion. This research was supported by grants from the National Natural Science Foundation of China (grant numbers: 81672389, 81672647, 81702954, and 81372214), Key Science and Technology Program of Anhui Province (grant number: 1704a0802167), Natural Science Research Project of Anhui Province (grant number: KJ2016A329), Science and Technology Service Network Initiative of Chinese Academy of Sciences (grant number: KFJ-STS-SCYD-010), The Major/Innovative Program of Development Foundation of Hefei Center for Physical Science and Technology (grant number: 2016FXCX006), Key Technologies R&D Programme of Anhui Province (grant number: 1604a0802103), and the 100-Talent Program of Chinese Academy of Sciences.
Disclosure
The authors report no conflicts of interest in this work.
References
Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. | ||
Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. | ||
Diefenbach RJ, Mackay JP, Armati PJ, Cunningham AL. The C-terminal region of the stalk domain of ubiquitous human kinesin heavy chain contains the binding site for kinesin light chain. Biochemistry. 1998;37(47):16663–16670. | ||
Hirokawa N. Kinesin and dynein superfamily proteins and the mechanism of organelle transport. Science. 1998;279(5350):519–526. | ||
Lawrence CJ, Dawe RK, Christie KR, et al. A standardized kinesin nomenclature. J Cell Biol. 2004;167(1):19–22. | ||
Miki H, Setou M, Kaneshiro K, Hirokawa N. All kinesin superfamily protein, KIF, genes in mouse and human. Proc Natl Acad Sci U S A. 2001;98(13):7004–7011. | ||
Echard A, Jollivet F, Martinez O, et al. Interaction of a Golgi-associated kinesin-like protein with Rab6. Science. 1998;279(5350):580–585. | ||
Verhey KJ, Hammond JW. Traffic control: regulation of kinesin motors. Nat Rev Mol Cell Biol. 2009;10(11):765–777. | ||
Taniuchi K, Nakagawa H, Nakamura T, et al. Down-regulation of RAB6KIFL/KIF20A, a kinesin involved with membrane trafficking of discs large homologue 5, can attenuate growth of pancreatic cancer cell. Cancer Res. 2005;65(1):105–112. | ||
Gasnereau I, Boissan M, Margall-Ducos G, et al. KIF20A mRNA and its product MKlp2 are increased during hepatocyte proliferation and hepatocarcinogenesis. Am J Pathol. 2012;180(1):131–140. | ||
Claerhout S, Lim JY, Choi W, et al. Gene expression signature analysis identifies vorinostat as a candidate therapy for gastric cancer. PLoS One. 2011;6(9):e24662. | ||
Yan GR, Zou FY, Dang BL, et al. Genistein-induced mitotic arrest of gastric cancer cells by downregulating KIF20A, a proteomics study. Proteomics. 2012;12(14):2391–2399. | ||
Marin-Acevedo JA, Dholaria B, Soyano AE, et al. Next generation of immune checkpoint therapy in cancer: new developments and challenges. J Hematol Oncol. 2018;11(1):39. | ||
Sundar R, Rha SY, Yamaue H, et al. A Phase I/Ib study of OTSGC-A24 combined peptide vaccine in advanced gastric cancer. BMC Cancer. 2018;18(1):332. | ||
Fujiwara Y, Sugimura K, Miyata H, et al. A Pilot study of post-operative adjuvant vaccine for advanced gastric cancer. Yonago Acta Med. 2017;60(2):101–105. | ||
Fujiwara Y, Okada K, Omori T, et al. Multiple therapeutic peptide vaccines for patients with advanced gastric cancer. Int J Oncol. 2017;50(5):1655–1662. | ||
Tatsuta M, Iishi H, Baba M, Yano H, Uehara H, Nakaizumi A. Attenuation by genistein of sodium-chloride-enhanced gastric carcinogenesis induced by N-methyl-N’-nitro-N-nitrosoguanidine in Wistar rats. Int J Cancer. 1999;80(3):396–399. | ||
Zhou HB, Chen JJ, Wang WX, Cai JT, Du Q. Apoptosis of human primary gastric carcinoma cells induced by genistein. World J Gastroenterol. 2004;10(12):1822–1825. | ||
Li YS, Wu LP, Li KH, et al. Involvement of nuclear factor κB (NF-κB) in the downregulation of cyclooxygenase-2 (COX-2) by genistein in gastric cancer cells. J Int Med Res. 2011;39(6):2141–2150. | ||
Yanagihara K, Takigahira M, Mihara K, et al. Inhibitory effects of isoflavones on tumor growth and cachexia in newly established cachectic mouse models carrying human stomach cancers. Nutr Cancer. 2013;65(4):578–589. | ||
Mai Z, Blackburn GL, Zhou JR. Genistein sensitizes inhibitory effect of tamoxifen on the growth of estrogen receptor-positive and HER2-overexpressing human breast cancer cells. Mol Carcinog. 2007;46(7):534–542. | ||
Prete SP, Rossi L, Correale PP, et al. Combined effects of protein kinase inhibitors and 5-fluorouracil on CEA expression in human colon cancer cells. Pharmacol Res. 2005;52(2):167–173. | ||
Hu XJ, Xie MY, Kluxen FM, Diel P. Genistein modulates the anti-tumor activity of cisplatin in MCF-7 breast and HT-29 colon cancer cells. Arch Toxicol. 2014;88(3):625–635. | ||
Meng D, Chen Y, Yun D, et al. High expression of N-myc (and STAT) interactor predicts poor prognosis and promotes tumor growth in human glioblastoma. Oncotarget. 2015;6(7):4901–4919. | ||
Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6(1):1–6. | ||
Lánczky A, Nagy Á, Bottai G, et al. miRpower: a web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients. Breast Cancer Res Treat. 2016;160(3):439–446. | ||
Cui J, Chen Y, Chou WC, et al. An integrated transcriptomic and computational analysis for biomarker identification in gastric cancer. Nucleic Acids Res. 2011;39(4):1197–1207. | ||
D’Errico M, de Rinaldis E, Blasi MF, et al. Genome-wide expression profile of sporadic gastric cancers with microsatellite instability. Eur J Cancer. 2009;45(3):461–469. | ||
Cho JY, Lim JY, Cheong JH, et al. Gene expression signature-based prognostic risk score in gastric cancer. Clin Cancer Res. 2011;17(7):1850–1857. | ||
Neef R, Grüneberg U, Barr FA. Assay and functional properties of Rabkinesin-6/Rab6-KIFL/MKlp2 in cytokinesis. Methods Enzymol. 2005;403:618–628. | ||
Nguyen PA, Groen AC, Loose M, et al. Spatial organization of cytokinesis signaling reconstituted in a cell-free system. Science. 2014;346(6206):244–247. | ||
Neef R, Preisinger C, Sutcliffe J, et al. Phosphorylation of mitotic kinesin-like protein 2 by polo-like kinase 1 is required for cytokinesis. J Cell Biol. 2003;162(5):863–876. | ||
Tcherniuk S, Skoufias DA, Labriere C, et al. Relocation of AuroraB and survivin from centromeres to the central spindle impaired by a kinesin-specific MKLP-2 inhibitor. Angew Chem Int Ed Engl. 2010;49(44):8228–8231. | ||
Gruneberg U, Neef R, Honda R, Nigg EA, Barr FA. Relocation of Aurora B from centromeres to the central spindle at the metaphase to anaphase transition requires MKlp2. J Cell Biol. 2004;166(2):167–172. | ||
Adams RR, Carmena M, Earnshaw WC. Chromosomal passengers and the (aurora) ABCs of mitosis. Trends Cell Biol. 2001;11(2):49–54. | ||
Ruchaud S, Carmena M, Earnshaw WC. Chromosomal passengers: conducting cell division. Nat Rev Mol Cell Biol. 2007;8(10):798–812. | ||
D’Avino PP, Savoian MS, Glover DM. Cleavage furrow formation and ingression during animal cytokinesis: a microtubule legacy. J Cell Sci. 2005;118(Pt 8):1549–1558. | ||
Imai K, Hirata S, Irie A, et al. Identification of HLA-A2-restricted CTL epitopes of a novel tumour-associated antigen, KIF20A, overexpressed in pancreatic cancer. Br J Cancer. 2011;104(2):300–307. | ||
Spagnuolo C, Russo GL, Orhan IE, et al. Genistein and cancer: current status, challenges, and future directions. Adv Nutr. 2015;6(4):408–419. | ||
Satoh H, Nishikawa K, Suzuki K, et al. Genistein, a soy isoflavone, enhances necrotic-like cell death in a breast cancer cell treated with a chemotherapeutic agent. Res Commun Mol Pathol Pharmacol. 2003;114:113149–158. | ||
Suzuki R, Kang Y, Li X, et al. Genistein potentiates the antitumor effect of 5-fluorouracil by inducing apoptosis and autophagy in human pancreatic cancer cells. Anticancer Res. 2014;34(9):4685–4692. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.