Back to Journals » OncoTargets and Therapy » Volume 12
Upregulation of hypoxia-inducible factor 1α mRNA expression was associated with poor prognosis in patients with hepatocellular carcinoma
Authors Cheng W , Cheng Z, Yang Z , Xing D, Zhang M
Received 5 December 2018
Accepted for publication 21 July 2019
Published 9 August 2019 Volume 2019:12 Pages 6285—6296
DOI https://doi.org/10.2147/OTT.S197077
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Jianmin Xu
Wei Cheng,1 Ziwei Cheng,1 Zongguo Yang,2 Dongwei Xing,1 Minguang Zhang1
1Department of Radiology, Shanghai Municipal Hospital of Traditional Chinese Medicine, Shanghai University of Traditional Chinese Medicine, Shanghai 200071, People’s Republic of China; 2Department of Integrative Medicine, Shanghai Public Health Clinical Center, Fudan University, Shanghai 201508, People’s Republic of China
Background: HIF1α mRNA expression in hepatocellular carcinoma (HCC) tissues and its relationship with the prognosis in HCC patients is still unclear. We performed this study to investigate the expression of HIF1α mRNA and its correlation with the prognosis in HCC patients.
Materials and methods: GSE14520 and Oncomine database were used to analyse the differential expression of HIF1α mRNA among HCC tissues and corresponding peritumour tissues or normal liver tissues. The relationship between HIF1α mRNA expression and the clinicopathological features and survival in HCC patients was analysed using the GSE14520 dataset. CCK-8 assay, wound-healing assay, transwell invasion assay, tube formation assay, and subcutaneous xenograft tumour assays using nude mice were used to confirm the function of HIF1α.
Results: Expression of HIF1α mRNA was significantly upregulated in HCC tissues (P<0.05 in all cases); this was supported by the results of the Western blotting (P=0.031) and IHC analyses. Our analysis of the clinicopathological features of HCC patients indicated that high HIF1α mRNA expression was strongly related with TNM stage III (P=0.002) and BCLC stage C (P=0.038). Survival analysis demonstrated that HCC patients with high HIF1α mRNA expression had a short overall survival (OS) (P=0.048), but showed no significant difference in recurrence-free survival (RFS) (P=0.066) compared to patients with low HIF1α mRNA expression. We further demonstrated that HIF1α promoted the proliferation, migration, invasion, and angiogenic ability of HCC cells, by using the stably transformed SK-Hep1 and Hep-3B cell lines showing HIF1α overexpression. Finally, xenograft tumour models of nude mice showed that RNA interference-mediated HIF1α silencing suppressed tumour growth and angiogenesis in HCC.
Conclusion: Our study suggests that the upregulation of HIF1α mRNA, which is found in HCC tissues and associated with poor prognosis in HCC patients, contributed to the proliferation, migration, invasion, and angiogenic ability of HCC cells.
Keywords: HIF1α, mRNA, prognosis, hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC) is the fifth most common tumour and the second major cause of tumour-associated deaths worldwide.1,2 More than half of all HCC patients in the world have been diagnosed and treated in China.3 Liver tumour resection and liver transplantation are the major treatment strategies for early HCC. However, treatment of advanced HCC mainly depends on transcatheter arterial chemoembolisation, radiotherapy, chemotherapy, and targeted therapy with sorafenib.4–6 Most HCC patients have been diagnosed at the advanced stage of the disease, especially in China, and have lost the chance of surgery and transplantation, with relatively short overall survival (OS) and recurrence-free survival (RFS).
Migration and invasion are the main causes of cancer progression and treatment failure in advanced-stage HCC patients.7 Angiogenesis also plays a vital role in the development of HCC, which is a highly vascularised solid tumour.8 Thus, suppression of HCC cell migration and invasion, and angiogenesis is the current trend of HCC treatment research.
Hypoxia-inducible factor 1α (HIF1α) was discovered by Semenza in 1992 during the analysis of the expression of the human erythropoietin gene.9 HIF1α is a heterodimer protein comprising an oxygen-sensitive α subunit and constantly expressed β subunit.10,11 HIF1α plays a critical role in regulating cellular responses to hypoxia and the biological behaviour of tumours by affecting cell apoptosis, cell viability, vasomotor function, and energy metabolism.12–14 HIF1α has also been reported to promote HCC cells migration and invasion, and angiogenesis by activating the expression of downstream-related genes such as VEGF.15–19 Thus, elucidating the mechanisms underlying the effects of HIF1α in HCC is urgently needed for the prevention and treatment of liver cancer.
In recent years, many articles have reported that the overexpression of the HIF1α protein is associated with poor OS and RFS in HCC patients.20–25 However, very few studies have reported the correlation between HIF1α mRNA overexpression and prognosis of liver cancers.26,27 The relationship between the upregulation of HIF1α mRNA expression and prognosis of HCC patients is still unclear. It is very essential to explore the HIF1α mRNA expression and its prognostic value and specific function in HCC.
This study aims to explore the expression of HIF1α mRNA in HCC tissues, and its relationship with the prognosis in HCC patients using GSE14520 and two datasets from the Oncomine database. We also verified the function of HIF1α by constructing the stably transformed SK-Hep1 and Hep-3B cell lines showing HIF1α overexpression and HIF1α silencing (by RNA interference).
Materials and methods
Patients and ethics statement
HCC tissues and corresponding peritumour tissues were obtained from 12 patients admitted at the Shanghai Public Health Clinical Center, Fudan University. These HCC patients were diagnosed according to histopathological analysis.28 Seven patients were men and five were women; the patients were between 25 and 70 years old. Among these patients, three, four, and five patients showed TNM stage IA, IB, and II HCC, respectively. No anti-tumour treatment was administered to these patients before operation. They all signed the informed consent forms during their hospitalisation. The protocols for this study and informed consent documents were reviewed and approved by the Ethics Committee of the Shanghai Public Health Clinical Center, Fudan University. This study was conducted in accordance with the Declaration of Helsinki.
Datasets source
The Roessler Liver and Roessler Liver 2 datasets were chosen from the Oncomine database (https://www.oncomine.org).29 The GSE14520 dataset, which contains data for 242 HCC patients and the related information regarding their clinicopathological features, was downloaded from Gene Expression Omnibus (GEO) database in the National Center for Biotechnology Information (NCBI). The expression of HIF1α mRNA in HCC tissues was compared with that in the corresponding peritumour tissues or normal liver tissues. The immunohistochemistry (IHC) data of the HIF1α protein in the HCC tissues and normal liver tissues were downloaded from The Human Protein Atlas (http://www.proteinatlas.org).
Materials and reagents
Dulbecco’s modified Eagle’s medium (DMEM) and penicillin/streptomycin were purchased from Hyclone (Logan, USA). Foetal bovine serum (FBS) was purchased from Gibco (South America). Anti-HIF1α monoclonal antibodies were purchased from CST (Boston, USA). Anti-VEGF monoclonal antibodies were purchased from RD (Minneapolis, USA). Horseradish peroxidase (HRP)-conjugated secondary antibodies, New Super ECL Assay kit, and anti-β-actin antibodies were purchased from KeyGEN BioTECH (Nanjing, China). Transwell chambers and Matrigel were purchased from Corning Life Sciences (8-μm pores, Tewksbury, MA, USA) and BD Biosciences (San Jose, CA, USA), respectively. The QRT-PCR kit was purchased from Takara (Shiga, Japan). The Cell Counting kit-8 (CCK-8) assay was purchased from Dojindo Molecular Technologies, Inc. The BCA Protein Assay Kit was purchased from Beyotime Biotechnology (Shanghai, China). Recombinant lentiviral expression vectors (LV-HIF1α, LV-Con, LV-ShHIF1α, and LV-Scramble), and polybrene were obtained from Genechem (Shanghai, China). BALB/c nude mice were purchased from Shanghai Jiesijie Experimental Animal Co., Ltd (Shanghai, China).
Cell culture
Human umbilical vein endothelial cells (HUVECs) and two types HCC cell lines (SK-Hep1 and Hep-3B) were purchased from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China), and then cultured in 5% CO2 at 37 °C in DMEM containing 10% FBS and 1% penicillin-streptomycin. A hypoxic culture was defined when the cells were cultured under conditions of 5% CO2, 94% N2, and 1% O2 at 37 °C in DMEM with 10% FBS and 1% penicillin-streptomycin.
In vitro transfection
LV-HIF1α and LV-ShHIF1α (sequence: CATCTCCATCTCCTACCCACAT) are lentiviral vectors that can express GFP, and also overexpress and silence HIF1α, respectively. LV-Con and LV-Scramble (sequence: TTCTCCGAACGTGTCACGT) are also lentiviral vectors that only express GFP, without specific HIF1α-overexpression or -silencing (via RNA interference) effects; cells transfected with these constructs have been commonly used as controls. For lentivirus infection, 2×105 cells/well were seeded into six-well culture plates for 24 h. Further, 10 µl of LV-HIF1α (2×108 TU/ml), 2.5 µl of LV-Con (8×108 TU/ml), 5 µl of LV-ShHIF1α (4×108 TU/ml), or 2 µl of LV-Scramble (1×109 TU/ml) and 1 µl of polybrene (10 mg/ml) were added to 1.0 ml of DMEM containing 10% FBS without antibiotics. The old culture medium was replaced with the mixture containing the lentivirus expression vectors, polybrene, and DMEM without antibiotics. After 12 h, the cell culture medium was replaced again with 2 ml of DMEM containing 10% FBS and antibiotics. After 72 h of incubation, the fluorescence of the lentivirus-infected cells was observed using fluorescence microscopy. The infected cells can be cultured continually or collected immediately and used for QRT-PCR assay or Western blotting.
QRT-PCR assay
Total RNA from the cultured cells was extracted using TRIzol reagent, according to the manufacturer’s instructions. A QRT-PCR kit and 0.5 µg of total RNA were used to perform the QRT-PCR analysis. All the primers used were synthesised by Sangon Biotech. The specific primers used are listed in Table 1.
 |
Table 1 Primers used for QRT-PCR |
Western blotting analysis
Cells were scraped off from the six-well culture plates and lysed using RIPA lysis buffer containing protease inhibitors and phenylmethylsulfonyl fluoride (PMSF) for 30 min on ice. The protein concentrations were quantified using the BCA Assay Kit. Equal amounts of protein (50–100 μg) and the corresponding amounts of 5× loading buffer were mixed, and these mixtures were subjected to 7.5% or 12% sodium dodecyl sulphate-polyacrylamide gel electrophoresis; the protein bands were then transferred onto pure nitrocellulose blotting membranes. After blocking the membranes with 5% non-fat milk at room temperature for 2 h, the membranes were incubated overnight at 4 °C with the primary antibodies (diluted at 1:1000), and then with the HRP-conjugated secondary antibodies (diluted at 1:2000) at room temperature for 1 h. The targeted proteins were visualised using a New Super ECL Assay kit and then exposed onto films for visualisation.
Determination of cell viability by the CCK-8 assay
Cell viabilities were assessed with the Cell Counting Kit-8 assay. A total of 10000 cells/well were plated onto 96-well culture plates and then cultured for 24 h. The cells were washed thrice with sterile PBS; then, 10 µl of CCK-8 regent and 90 µl of fresh medium was added to the cells. Finally, the cells were incubated for 1 h at 37 °C. The optical density (OD) of the samples was measured at 450 nm using a microplate reader. All measurements were carried out in triplicate. We calculated the cell viability index based on the following formula: (experimental optical density value/control optical density value) ×100%.30
Wound healing assay
Cells were plated in six-well culture plates. When the cells grew to 80–90% confluence, the cell layers were scratched in the form of a straight line (in the middle of each well) using a sterile white pipette tip. The cells were then rinsed thrice with fresh serum-free DMEM to wash away the cell debris; these cells were cultured for 24 h. The scratched cell layers were photographed with an inverted microscope at 0 h and 24 h. These experiments were repeated thrice.
Transwell invasion assay
We performed the transwell assay to detect the invasive ability of the cells. First, 600 µl of DMEM containing 20% FBS was dispensed into the lower chamber. Next, 100 µl of Matrigel diluted with fresh serum-free DMEM (in a ratio of 1:3) was dispensed into the upper chamber and allowed to solidify for 30 min at 37 °C before the cells were plated. The cells were then plated into the upper chamber, followed by incubation for 72 h. The invasive cells were analysed by crystal violet staining; the number of invasive cells was counted under a light microscope. These experiments were repeated thrice.
HUVECS tube formation assay
Human HCC cells infected with the lentiviral constructs (LV- HIF1α and LV-Con) were seeded into six-well plates and cultured for 24 h; then, the cell supernatants were collected as conditional medium for subsequent use. Next, 50 µl of Matrigel was dispensed into the 96-well plates and allowed to solidify at 37 °C for 30 min, after which 100 µl of conditional medium was added to the 96-well plates containing Matrigel. HUVECs (2.5×104 cells) were seeded onto the Matrigel, followed by incubation at 37 °C for 24 h. The formation of capillary-like structures was photographed, and the tube numbers were analysed using the Image J software. These experiments were repeated thrice.
In vivo xenograft model assay
In order to verify the function of HIF1α, we performed the in vivo xenograft model assay. Ten male BALB/c nude mice (aged four–six weeks old) were divided into two groups (LV-Scramble and LV-ShHIF1α groups). Further, 5×106 transformed Sk-Hep1 cells (transfected with LV-Scramble-Sk-Hep1 or LV-ShHIF1α-Sk-Hep1) (in a volume of 100 µl) were subcutaneously injected into the right flank of the nude mice after one week of adaptive feeding. The size of the xenograft tumours was measured every seven days for five weeks and calculated using the formula V =0.5×L×W2 (V, volume; L, length; W, width). All animals were sacrificed by euthanasia and the xenograft tumours were obtained for the IHC assay.
IHC staining
Paraffin sections were deparaffinised in an oven and then hydrated. The sections were then treated with 3% H2O2 to block endogenous peroxidase, and were then subjected to thermal repair. After cooling, the sections were blocked with 10% normal goat serum for 1 h and then incubated with the primary antibodies (diluted at 1:200) against HIF1α and VEGF overnight at 4 °C. The sections were then incubated with HRP-conjugated secondary antibodies (diluted at 1:600) at room temperature for 1 h. Finally, all sections were counterstained with Diaminobenzidine and Mayer’s haematoxylin.
Statistical analysis
Categorical data were compared using the Chi-square test. Probabilities of OS and RFS were calculated using the Kaplan-Meier method, and the log-rank test was used to evaluate the differences between survival distributions. All experimental data were presented at the means ± standard deviations, and the significance of data from two groups was estimated using the Student’s t test. Statistical analyses were carried out using the IBM SPSS Statistics 21 software. P<0.05 was considered statistically significant for all the tests.
Results
Expression of HIF1α is significantly upregulated in HCC tissues
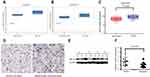
The ‘Roessler Liver’ dataset, which contains data for 22 HCC tissues and 21 normal liver tissues, and the ‘Roessler Liver 2ʹ dataset, which contains data for 225 HCC tissues and 220 normal liver tissues, are from the Oncomine database. The GSE14520 dataset contains data for 242 HCC tissues and 239 peritumour tissues. Analysis of these datasets showed that the expression of HIF1α mRNA was significantly upregulated in HCC tissues, compared to the case in normal liver tissues or corresponding peritumour tissues (P<0.001, Figure 1A; P<0.001, Figure 1B; and P=0.031, Figure 1C). IHC results from the human protein atlas database demonstrated that a positive staining of the HIF1α protein was observed in HCC tissues, whereas the normal liver tissues showed negative staining for the HIF1α protein (Figure 1D). Finally, based on the Western blotting analysis, HIF1α protein expression was also observed to be upregulated in HCC tissues, compared to the case for the corresponding peritumour tissues (P=0.031, Figure 1E and F).
Clinicopathological characteristics of patients and their relationship with the HIF1α mRNA level in HCC
Clinicopathological characteristics of all HCC patients from the GSE14520 dataset are fully described in Table 2. This dataset comprises data for 242 HCC patients, with complete survival information. Our study included 193 HCC patients younger than 60 years old. 210 HCC patients were men. With regards to the HBV status, the numbers of HCC patients described as chronic carriers (CCs), active viral replication-chronic carriers (AVR-CCs), and as those with unavailable HBV status were 162, 57, and 6, respectively. The number of HCC patients with an ALT level of more than 50 U/L was 100. The number of HCC patients with an AFP level of more than 300 ng/ml was 97. The number of HCC patients with multinodular tumours was 51. The number of HCC patients with a main tumour size of more than 5 cm was 88. The number of cirrhosis patients was 223. The numbers of patients with TNM stage I, II, and III HCC were 97, 78, and 59, respectively, and with BCLC stage A, B, and C were 153, 24, and 27, respectively. Based on the median HIF1α mRNA expression levels, the patients were divided into the high expression and low expression groups. HCC patients with an HIF1α mRNA expression ≥ the median HIF1α mRNA expression level were considered as those with high HIF1α expression, while the others were considered as those with low HIF1α expression. There was no significant difference between the high HIF1α mRNA expression group and low HIF1α mRNA expression group when comparing age (≥60 versus <60), gender (male versus female), HBV status (CC and AVR-CC versus NA), ALT (>50 U/L versus ≤50 U/L), AFP (>300 ng/mL versus ≤300 ng/mL), multinodular tumours (Yes versus No), main tumour size (>5 cm versus ≤5 cm), and cirrhosis (Yes versus No). However, HCC patients with high HIF1α mRNA expression had a higher tendency to be associated with TNM stage III and BCLC stage C of the disease than those with low HIF1α mRNA expression (P=0.002 and P=0.038, respectively; Table 2). Thus, the upregulation of HIF1α mRNA expression was found to be correlated with TNM stage III and BCLC stage C of liver cancer.
 |
Table 2 Correlations between HIF1α mRNA expression and the clinic-pathologic variables of HCC patients |
Overexpression of HIF1α mRNA contributes to poor prognosis in HCC patients
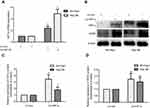
Kaplan-Meier analysis of data from the GSE14520 cohort demonstrated that HCC patients with high HIF1α mRNA expression showed poor OS, compared to those with low HIF-1α mRNA expression (HR=1.503, 95% CI=1.001–2.245, P=0.048, Figure 2A). There was no significant difference between the RFS of patients showing high and low HIF1α mRNA expression; however, with regards to the RFS, there was an obvious survival advantage for HCC patients with low HIF1α mRNA expression (HR=1.374, 95% CI=0.976–1.922, P=0.066, Figure 2B).
Establishment of the SK-Hep1 and Hep-3B cell lines stably transfected with the HIF1α gene (HIF1α overexpression)
In order to further explore the function of HIF1α in HCC, we constructed the stably transformed SK-Hep1 and Hep-3B cell lines with HIF1α overexpression using lentiviral vectors. Our results showed that HIF1α was significantly upregulated at both the mRNA and protein levels in SK-Hep1 and Hep-3B cells from the LV-HIF1α group, compared to the case for those from the LV-Con group in (Figure 3A–C). Additionally, we found that VEGF, a protein present downstream of HIF1α, was also upregulated in cells from the LV-HIF1α group (Figure 3B and D). Thus, we successfully constructed the stably transformed SK-Hep1 and Hep-3B cell lines with HIF1α overexpression.
Overexpression of HIF-1α promotes the proliferation, migration, invasion, and angiogenic ability of SK-Hep1 and Hep-3B cells
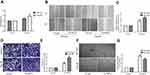
We performed the CCK-8 assay to assess the effect of HIF1α overexpression on the proliferation of SK-Hep1 and Hep-3B cells. Our results showed that the viability of SK-Hep1 and Hep-3B cells from the LV-HIF1α group increased significantly, compared to the case for those from the LV-Con group (Figure 4A). To identify the effects of HIF1α overexpression on the migration and invasion abilities of SK-Hep1 and Hep-3B cells, we performed the wound healing assay and transwell invasion assay. The results indicated that HIF1α overexpression significantly promoted the migration (Figure 4B and C) and invasion (Figure 4D and E) of SK-Hep1 and Hep-3B cells. Finally, our results also demonstrated that the SK-Hep1 and Hep-3B cell culture supernatants from the LV-HIF1α group significantly promoted the angiogenesis of HUVECs, compared to those from the LV-Con group (Figure 4F and G).
Establishment of the stably transformed SK-Hep1 and Hep-3B cell lines with HIF1α silencing using RNA interference, and the inhibitory effects of HIF1α silencing on HCC cell proliferation
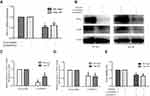
We constructed the stably transformed SK-Hep1 and Hep-3B cell lines by means of RNA interference-mediated HIF1α gene silencing to verify the effects of HIF1α on HCC cell proliferation, given the low expression of HIF1α in Hep-3B cells under conditions of normoxia. Our results showed that HIF1α was significantly downregulated at both the mRNA and protein levels in SK-Hep1 and Hep-3B cells from the LV-ShHIF1α group, compared to the case for those from the LV-Scramble group (Figure 5A–C). Additionally, we also found that the expression of VEGF, a protein present downstream of HIF1α, was downregulated in cells from the LV-ShHIF1α group (Figure 5B and D). Thus, we successfully constructed the stably transformed SK-Hep1 and Hep-3B cell lines showing HIF1α silencing (by RNA interference). We performed the CCK-8 assay to examine the effects of HIF1α silencing on the proliferation of SK-Hep1 and Hep-3B cells. Our results indicated that the viability of SK-Hep1 and Hep-3B cells from the LV-ShHIF1α group decreased significantly, compared to the case for those from the LV-Scramble group (Figure 5E).
RNA interference-mediated HIF1α silencing suppressed the growth of subcutaneous xenograft tumours in nude mice
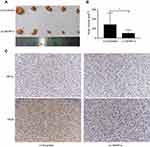
In order to further illustrate the function of HIF1α, we constructed xenograft models using BALB/c nude mice. Stably transformed Sk-Hep1 cells (LV-Scramble-Sk-Hep1 or LV-ShHIF1α-Sk-Hep1) were injected subcutaneously into the right flank of the nude mice. The size of the xenograft tumours was measured every seven days for five weeks. The results indicated that the xenograft tumours from mice injected with the LV-Scramble-transfected cells were significantly larger than those from mice injected with the LV-ShHIF1α-transfected cells (Figure 6A and B). The IHC results demonstrated that staining of the HIF1α and VEGF proteins in xenograft tumours from mice injected with the LV-Scramble-transfected cells was significantly stronger than in those from mice injected with the LV-ShHIF1α-transfected cells (Figure 6C).
Discussion
HCC is a malignant cancer with high morbidity and mortality. Although there are a large number of treatment strategies for HCC, the therapeutic effects are still not satisfactory. Thus, there is an urgent need to explore the new mechanisms underlying the invasion and metastasis of HCC and look for new predictive markers and targets for therapeutic interventions to improve the outcome of HCC treatment strategies.
HIF1α is a heterodimeric transcription factor containing the oxygen-sensitive α subunit and the constitutively expressed β subunit. Recent studies have demonstrated that the overexpression of HIF1α at the protein level is always linked with poor prognosis. Nevertheless, there are very few studies examining the correlation between the overexpression of HIF1α mRNA and its prognostic value in HCC patients. Hence, it is essential to clarify the effects of the upregulation of HIF1α mRNA expression on the pathological features and prognostic value in HCC patients, and the specific function of HIF1α in human HCC cell lines.
Our study outcomes showed that HIF1α mRNA expression was significantly upregulated in HCC tissues, compared to the case in normal liver tissues or corresponding peritumour tissues; these results were further supported by those of the Western blotting and IHC analyses. Results of the survival analysis demonstrated that HCC patients with high HIF1α mRNA expression showed a shorter OS, but not RFS, than HCC patients with low HIF1α mRNA expression. Additionally, we found that in HCC patients, high HIF1α mRNA expression was associated with TNM stage III and BCLC stage C of the disease, compared to the case for low HIF1α mRNA expression. In order to explore the function of HIF1α in detail, we successfully constructed the stably transformed SK-Hep1 and Hep-3B cell lines with HIF1α overexpression and HIF1α inhibition (by means of RNA interference to silence HIF1α expression) using lentiviral vectors. Results indicated that HIF1α overexpression significantly promoted the proliferation, migration, invasion, and angiogenic ability of HCC cells, and HIF1α inhibition significantly suppressed the proliferation of HCC cells. Finally, the results of the xenograft tumour assay using nude mice showed that HIF1α silencing suppressed the growth of xenograft tumours and the expression of the VEGF protein.
Considering the poor prognostic value of HIF1α in HCC patients, and that it promotes the proliferation, migration, invasion, and angiogenic ability of HCC cells, many researchers have been encouraged to deplete HIF1α mRNA from HCC patients to decrease the activity of tumour cells, with the aim of controlling the growth of HCC tumours. In addition, we may need to consider both the HCC patients’ pathological conditions and their HIF1α mRNA levels when formulating treatment strategies. HCC patients with high HIF1α mRNA levels may need more aggressive treatments after surgery. However, our study showed that there was no significant difference in the RFS of HCC patients showing high and low HIF1α mRNA expression. The reason for this may be the lack of a sufficient sample size. Hence, additional prospective investigations with larger sample sizes are still needed to confirm the exact prognostic value of HIF1α mRNA expression in HCC patients.
Our study has a few limitations. Firstly, in our research, the analysis of HIF1α overexpression and its prognostic value is restricted to the mRNA level, which may not necessarily reflect the function of HIF1α when it is activated. Secondly, our survival analysis mainly was only based on the GEO database; no other related survival data were considered. Thirdly, our results need to be verified in other HCC cell lines, such as the Huh7, bel7404, and SMMC-7721 cell lines.
Conclusion
Our study demonstrated that the upregulation of HIF1α mRNA expression was found in HCC tissues; it was associated with unfavourable OS and linked with TNM stage III and BCLC stage C in HCC patients. We also showed that HIF1α promoted the proliferation, migration, invasion, and angiogenic ability of HCC cells.
Acknowledgments
This work was supported by National Natural Science Foundation of China (Research Grants no. 81673743).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi:10.3322/caac.21387
2. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi:10.3322/caac.21262
3. Chen W, Zheng R, Baade PD, et al. Cancer Statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi:10.3322/caac.21338
4. Ni JY, Liu SS, Sun HL, et al. Transcatheter hepatic arterial infusion chemotherapy vs sorafenib in the treatment of patients with hepatocellular carcinoma of Barcelona clinic liver cancer stage C: a meta-analysis of Asian population. Onco Targets Ther. 2018;11:7883–7894. doi:10.2147/OTT.S156844
5. Qin S, Bai Y, Lim HY, et al. Randomized, multicenter, open-label study of oxaliplatin plus fluorouracil/leucovorin versus doxorubicin as palliative chemotherapy in patients with advanced hepatocellular carcinoma from Asia. J Clin Oncol. 2013;31(28):3501–3508. doi:10.1200/JCO.2012.44.5643
6. Yu Y, Feng M. Radiotherapy for hepatocellular carcinoma. Semin Radiat Oncol. 2018;28(4):277–287. doi:10.1016/j.semradonc.2018.06.005
7. Antonio C, Ramona S, Alessandro C, et al. Prognostic factors for hepatocellular carcinoma recurrence. World J Gastroentero. 2014;20(20):5935–5950. doi:10.3748/wjg.v20.i20.5935
8. Li S, Yao D, Wang L, et al. Expression characteristics of hypoxia-inducible factor-1α and its clinical values in diagnosis and prognosis of hepatocellular carcinoma. Hepat Mon. 2011;11(10):36–43. doi:10.5812/kowsar.1735143X.771
9. Semenza GL, Wang GL. A nuclear factor induced by hypoxia via de novo protein synthesis binds to the human erythropoietin gene enhancer at a site required for transcriptional activation. Mol Cell Biol. 1992;12(12):5447–5454. doi:10.1128/mcb.12.12.5447
10. Wang GL, Semenza GL. Purification and characterization of hypoxia inducible factor 1. J Biol Chem. 1995;270(3):1230–1237. doi:10.1074/jbc.270.3.1230
11. Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci. 1995;92(12):5510–5514. doi:10.1073/pnas.92.12.5510
12. Chun YS, Kim MS, Park JW. Oxygen-dependent and -independent regulation of HIF-1alpha. J Korean Med Sci. 2002;17(5):581–588. doi:10.3346/jkms.2002.17.5.581
13. Fang J, Yan L, Shing Y, Moses MA. HIF-1alpha-mediated up-regulation of vascular endothelial growth factor, independent of basic fibroblast growth factor, is important in the switch to the angiogenic phenotype during early tumorigenesis. Cancer Res. 2001;61(15):5731–5735.
14. Raheja LF, Genetos DC, Wong A, Yellowley CE. Hypoxic regulation of mesenchymal stem cell migration: the role of RhoA and HIF-1alpha. Cell Biol Int. 2011;35(10):981–989. doi:10.1042/CBI20100733
15. Carbajo-Pescador S, Ordonez R, Benet M, et al. Inhibition of VEGF expression through blockade of Hif1a and STAT3 signaling mediates the anti-angiogenic effect of melatonin in HepG2 liver cancer cells. Br J Cancer. 2013;109(1):83–91. doi:10.1038/bjc.2013.285
16. Chen C, Liu R, Wang J, Yan Z, Qian S, Zhang W. RNAi knockdown of hypoxia-inducible factor-1a decreased the proliferation, migration, and invasion of hypoxic hepatocellular carcinoma cells. Cell Biochem Biophys. 2015;71(3):1–8. doi:10.1007/s12013-014-0390-x
17. Luo D, Wang Z, Wu J, Jiang C, Wu J. The role of hypoxia inducible factor-1 in hepatocellular carcinoma. Biomed Res Int. 2014;2014(3–4):409272. doi:10.1155/2014/409272
18. Chen C, Lou T. Hypoxia inducible factors in hepatocellular carcinoma. Oncotarget. 2017;8(28):46691–46703. doi:10.18632/oncotarget.17358
19. Lin D, Wu J. Hypoxia inducible factor in hepatocellular carcinoma: A therapeutic target. World J Gastroenterol. 2015;21(42):12171–12178. doi:10.3748/wjg.v21.i42.12171
20. Dai X, Pi G, Yang SL, Chen GG, Liu LP, Dong HH. Association of PD-L1 and HIF-1α coexpression with poor prognosis in hepatocellular carcinoma. Transl Oncol. 2018;11(2):559–566. doi:10.1016/j.tranon.2018.02.014
21. Wang D, Zhang X, Lu Y, Wang X, Zhu L. Hypoxia inducible factor 1α in hepatocellular carcinoma with cirrhosis: association with prognosis. Pathol Res Pract. 2018;214(12):1987–1992. doi:10.1016/j.prp.2018.09.007
22. Huang MS, Wang L, Chen JW, et al. Regulation of COX-2 expression and epithelial-to-mesenchymal transition by hypoxia-inducible factor-1α is associated with poor prognosis in hepatocellular carcinoma patients post TACE surgery. Int J Oncol. 2016;48(5):2144–2154. doi:10.3892/ijo.2016.3421
23. Zhou BJ, Liu XL, Zhang BM, et al. The expression of FAP in hepatocellular carcinoma cells is induced by hypoxia and correlates with poor clinical outcomes. J Cancer. 2018;9(18):3278–3286. doi:10.7150/jca.25775
24. Zheng SS, Chen XH, Yin X, Zhang BH. Prognostic significance of HIF-1a expression in hepatocellular carcinoma: a meta-analysis. Plose One. 2013;8(6):e65753. doi:10.1371/journal.pone.0065753
25. Cao S, Yang S, Wu C, Wang Y, Jiang J, Lu Z. Protein expression of hypoxia-inducible factor-1 alpha and hepatocellular carcinoma: A systematic review with meta-analysis. Clin Res Hepatol Gastroenterol. 2014;38(5):598–603. doi:10.1016/j.clinre.2014.04.004
26. Dai CX, Gao Q, Qiu QS, et al. Hypoxia-inducible factor-1 alpha, in association with inflammation, angiogenesis and MYC, is a critical prognostic factor in patients with HCC after surgery. BMC Cancer. 2009;9(418):1–11. doi:10.1186/1471-2407-9-418
27. Qin Y, Liu HJ, Li M, et al. Salidroside improves the hypoxic tumor microenvironment and reverses the drug resistance of platinum drugs via HIF-1α signaling pathway. EBioMedicine. 2018;38:25–36. doi:10.1016/j.ebiom.2018.10.069
28. Bruix J, Sherman M, Llovet JM, et al. Clinical management of hepatocellular carcinoma. Conclusions of the Barcelona-2000 EASL conference. European association for the study of the liver. J Hepatol. 2001;35(3):421–430.
29. Roessler S, Jia HL, Budhu A, et al. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Res. 2010;70(24):10202–10212. doi:10.1158/0008-5472
30. Wang J, Ma Y, Jiang H, et al. Overexpression of von Hippel-Lindau protein synergizes with doxorubicin to suppress hepatocellular carcinoma in mice. J Hepatol. 2011;55(2):359–368. doi:10.1016/j.jhep.2010.10.043
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.