Back to Journals » OncoTargets and Therapy » Volume 13
Upregulation of Cullin 4B Promotes Gastric Cancer and Predicts Poor Prognosis
Authors Wu P, Hu H, Li J, Gong W
Received 15 October 2019
Accepted for publication 10 January 2020
Published 11 February 2020 Volume 2020:13 Pages 1235—1243
DOI https://doi.org/10.2147/OTT.S234706
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Arseniy Yuzhalin
Ping Wu,1 Haolin Hu,2 Jinwen Li,1 Wei Gong1
1Department of Oncology, Xiangyang Central Hospital, Affiliated Hospital of Hubei University of Arts and Science, Xiangyang, 441021, People’s Republic of China; 2Department of Surgery, Xiangyang Central Hospital, Affiliated Hospital of Hubei University of Arts and Science, Xiangyang, 441021, People’s Republic of China
Correspondence: Haolin Hu
Department of Surgery, Xiangyang Central Hospital, Affiliated Hospital of Hubei University of Arts and Science, Jingzhou St 39, Xiangyang 441021, People’s Republic of China
Tel +86 138 7168 9148
Email [email protected]
Aim: Cullin 4B (CUL4B) is a member of the cullin ubiquitin-ligase family, which participates in proteolysis. Aberrant CUL4B expression has been shown in many malignancies. This study aimed to elucidate oncogenic role of CUL4B in gastric cancer (GC).
Methods: CUL4B expression in GC tissues was examined by RT-PCR and immunohistochemistry. The proliferation, invasion and tumorigenicity of GC cells with CUL4B overexpression or knockdown were evaluated.
Results: CUL4B expression significantly increased in GC tissues, and was correlated to UICC stage and differentiation of GC, as well as poor overall survival and disease-free survival. Both univariate and multivariate analysis identified CUL4B as an independent predictor for GC patient prognosis. In addition, CUL4B promoted GC cell proliferation and invasion in vitro and tumor formation in vivo.
Conclusion: CUL4B is overexpressed to promote GC development and progression. CUL4B is a promising prognostic marker and therapeutic target for GC.
Keywords: CUL4B, gastric cancer, biomarker, prognosis
Introduction
Gastric cancer (GC) is a leading cause of cancer-related deaths and is most common in China.1,2 Despite the developments in diagnosis and treatment, 5-year survival of GC patients is still poor.3,4 Increasing evidences indicate that abnormal gene expression is involved in GC initiation and progression.5–8 Therefore, the identification of novel genes involved in GC is of significance for the early detection and treatment of GC.
Cullin 4B (CUL4B) is a member of the CUL4 subfamily of Cullin RING E3 ligase.9 CUL4B plays an important role to regulate gene expression, DNA damage and cell cycle.10 Mechanistically, CUL4B directly interacts with damage specific DNA binding protein 1 (DDB1) and ring-box 1 (RBX1) by acting as a scaffold to assemble two independent E3 ligases known as CRL4BDCAF11 and CRL4BDCAF13, which then catalyze the ubiquitination and degradation of the substrate p21 and PTEN, respectively.11 Since both p21 and PTEN are tumor suppressors, the overexpression of CUL4B would lead to the downregulation of p21 and PTEN, and promote tumorigenesis.12–18 However, the role of CUL4B in the tumorigenesis of GC and prognostic value of CUL4B in GC remains unclear.
In the present study, we first detected the expression of CUL4B in GC, and then evaluated the correlation of CUL4B expression with clinicopathological parameters of GC patients. Furthermore, we investigated the role of CUL4B in GC by examining the effects of CUL4B overexpression and knockdown on the biological activities of GC cells in vitro and in vivo.
Materials and Methods
Patients
GC tissues were dissected from 50 GC patients (32 men and 18 women) who had surgery for GC and had not undergone radiotherapy or chemotherapy. The tumor grade and stage were judged according to the guidelines of UICC. Disease-free survival (DFS) and overall survival (OS) indicated the time from initial surgery to recurrence/metastasis and death, respectively. This study was approved by Ethics Committee of Hubei University of Arts and Science and all patients signed informed consent.
Immunohistochemistry
Tissue microarray (TMA) including 190 paired GC samples was purchased from Outdo Biotech (Shanghai, China). Tumor sections were dewaxed and rehydrated, and then incubated in 3% H2O2 for blocking endogenous peroxidase activity. Next, the sections were incubated in boiled citrate buffer (pH 6.0) for antigen retrieval. The sections were then incubated with CUL4B antibody (Abcam, Cambridge, UK) overnight at 4°C, followed by sequential incubation with secondary antibodies and diaminobenzidine (DAB). The sections were counterstained with hematoxylin and observed by two investigators independently in a blind manner. The intensity of staining was scored as 0 (no), 1 (mild), 2 (moderate), and 3 (strong). The area of staining was scored as 0 (0), 1 (1–25%), 2 (26–50%), 3 (51–75%), and 4 (76–100%). The staining score was the sum of the score of staining intensity and area and judged as negative (0–1), weak (2–4) and strong (5–6).
Real-Time PCR
Total RNA was prepared from tissues or cells using TRIzol (Invitrogen). cDNA was synthesized using First Strand cDNA Synthesis Kit (Fermentas, MA, USA). PCR was performed using cDNA, SYBR green (Takara, Shiga, Japan) and the following primers: CUL4B forward 5ʹ-CCTGGAGTTTGTAGGGTTTGAT-3ʹ, reverse 5ʹ-GAGACGGTGGTAGAAGATTTGG-3ʹ; Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) forward 5′-GGAGCGAGATCCCTCCAAAAT-3′, reverse 5′-GGCTGTTGTCATACTTCTCATGG-3′. The relative CUL4B mRNA level was normalized to GAPDH and calculated by 2−∆∆Ct method.
Western Blot Analysis
Total protein was extracted from tissues or cells using RIPA buffer (Beyotime, Jiangsu, China). Equal amounts of proteins were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred onto polyvinylidene fluoride (PVDF) membranes. The membranes were incubated with primary antibody for CUL4B (Abcam, Cambridge, UK) and β-actin (Santa Cruz, CA, USA) overnight at 4°C, followed by sequential incubation with secondary antibody and ECL reagent (Millipore, MA, USA).
Cell Transfection
Human GC cell lines BGC-823, AGS, HGC-27, MKN-28, MKN-45, SGC-7901, MGC-803 and normal gastric mucosa cell line GES-1 were purchased from Chinese Academy of Science, and cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS, Gibco, CA, USA) in a humidified atmosphere with 5% CO2 at 37°C. BGC-823 and MKN45 cells were cultured to around 70% confluency, and then were transfected with CUL4B siRNA and pcDNA3.1-CUL4B plasmid or corresponding controls (all from Biolink Biotech, Shanghai, China), respectively, by using Lipofectamine 2000 (Invitrogen, CA, USA). Culture medium was changed 24 h after transfection, the cells were cultured for another 24 hrs and then collected for following experiments.
Cell Proliferation Assay
The proliferation of BGC-823 and MKN45 cells was evaluated using cell counting kit-8 (CCK-8) assay (Dojindo, Kumamoto, Japan). In brief, cells were seeded in 96-well plates at 2000 cells/well and cultured for 24, 48, 72, 96, and 120 h. Then, the cells were incubated with 10 μL of CCK-8 solution at 37°C, and the absorbance at 450 nm was calculated.
Colony Formation Assay
Log-phase BGC-823 and MKN45 cells were seeded in 6-well plates and cultured for 14 days. Next, the cells were fixed, stained with Giemsa, and the colonies were counted under microscope.
Cell Invasion Assay
The invasion of BGC-823 and MKN45 cells was evaluated by using transwell chamber with Matrigel (Millipore, MA, USA) as described previously.19 In brief, 104 cells were seeded in the upper compartment of transwell chamber, and the bottom chamber was filled with medium supplemented with 10% FBS. After incubation for 48 h, cells that invaded to the bottom chamber were fixed and stained with crystal violet.
Nude Mice Xenograft Model
Animal experiments were approved by Animal Care and Use Committee of Hubei University of Arts and Science and performed in accordance with National Guideline for ethical review of animal welfare (GB/T 35892–2018, China). Four-week-old male BALB/C nude mice were divided randomly into two groups (n= 3), and 107 BGC-823/si-CUL4B and BGC-823/Scramble cells were injected subcutaneously into the left and right groin, respectively. Five weeks later, the mice were euthanized, and the tumor volume and mass were measured.
Statistical Analysis
All data were analyzed by SPSS 22.0 software (SPSS, Chicago, IL, USA). Differences in the groups were analyzed by Student’s t-test or Fisher’s exact test. Survival rate was calculated using Kaplan–Meier curve. The risk of individual factors was calculated by Cox proportional hazard model. P<0.05 was considered significant.
Results
Upregulation of CUL4B in GC
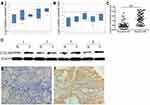
We first examined two independent GC datasets from the Oncomine database and observed significantly high expression of CUL4B in GC (Figure 1A and B). PCR analysis of fifty paired randomly selected GC specimens confirmed higher CUL4B mRNA levels in GC tissues compared to paired normal mucosa (Figure 1C). Moreover, Western blot analysis revealed higher CUL4B protein levels in GC tissues compared to paired normal mucosa (Figure 1D).
Association of CUL4B Expression with Clinical Features of GC
To investigate the association of CUL4B expression with clinical characteristics of GC, we performed CUL4B staining in TMA including 190 paired specimens. In normal mucosa, 116 (61.1%) showed negative CUL4B staining (Figure 1E), and only 20 (10.5%) showed strong CUL4B staining. In contrast, in GC tissues 92 (48.4%) showed strong CUL4B staining (Figure 1F), 66 (34.7%) showed weak CUL4B staining, and 32 (16.8%) showed negative CUL4B staining (Table 1).
 |
Table 1 CUL4B Expression in Normal Mucosa and GC Tissues |
The association of CUL4B expression and clinicopathologic parameters of GC was summarized in Table 2. Elevated CUL4B expression in GC was significantly associated with T classification (P<0.001), N classification (P<0.001), UICC stage (P<0.001) and tumor differentiation (P=0.007), but was not significantly associated with other clinicopathological parameters including gender, age, tumor size or location (P>0.05).
 |
Table 2 Association Between CUL4B Expression and Clinicopathological Features |
Prognostic Value of CUL4B in GC
Patients who were positive for CUL4B staining in GC tissues showed significantly worse DFS and OS compared to those who were negative for CUL4B staining in GC tissues (Figure 2), suggesting that elevated CUL4B expression predicts poor outcomes in these patients.
 |
Figure 2 Comparison of disease-free survival (P=0.013) and overall survival (P=0.017) in patients with CUL4B positive and CUL4B negative tumors. |
Based on univariate analysis, T classification, N classification, UICC stage, tumor differentiation and CUL4B were identified as significant independent prognostic factors. Based on multivariate analysis, CUL4B was identified as an independent prognostic factor for both DFS and OS (Tables 3 and 4).
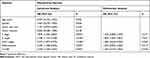 |
Table 3 Univariate and Multivariate Cox Proportional Hazard Models for Disease-Free Survival |
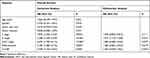 |
Table 4 Univariate and Multivariate Cox Proportional Hazard Models for Overall Survival |
CUL4B Promotes the Proliferation, Migration and Invasion of GC Cells in vitro
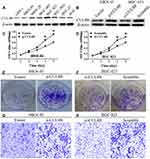
CUL4B protein expression levels in gastric mucosa cell line and GC cell lines were compared (Figure 3A). Among GC cell lines, MKN-45 cells had the lowest CUL4B expression and were used to generate CUL4B overexpression cell line, while BGC-823 cells had the highest CUL4B expression and were used to generate CUL4B knockdown cell line (Figure 3B).
CCK-8 assay showed that CUL4B knockdown or overexpression inhibited or promoted GC cell growth in vitro, respectively (Figure 3C and D). Moreover, clone formation assay demonstrated that CUL4B knockdown or overexpression decreased or increased GC cell colony formation, respectively (Figure 3E and F). Furthermore, transwell assays indicated that CUL4B knockdown or overexpression could decrease or increase GC cell invasion, respectively (Figure 3G and H). Collectively, these data suggest that CUL4B promotes GC cell proliferation and invasion.
CUL4B Knockdown Inhibits Xenograft Tumor in Nude Mice
To confirm the oncogenic role of CUL4B in GC, CUL4B knockdown and control BGC-823 cells were injected into the left and right groin of nude mice, respectively. CUL4B knockdown group generated smaller subcutaneous xenografts in nude mice compared with control group (Figure 4A–D). These results further support that CUL4B promotes GC tumor growth in vivo.
Discussion
CUL4B regulates a wide spectrum of biological processes such as DNA replication, DNA damage and cell cycle progression.20 Notably, CUL4B is crucially involved in tumor development and progression.15–18 Silencing CUL4B suppressed non-small cell lung cancer cell proliferation, invasion and epithelial mesenchymal transition (EMT) through canonical Wnt pathway.15 Furthermore, CUL4B knockdown significantly inhibited proliferation and induced apoptosis in osteosarcoma cells.18 In addition, abnormally elevated CUL4B level was correlated with differentiation, invasion, lymph node metastasis and advanced stage of colon cancer, predicting an unfavorable prognosis.16 In node-negative breast cancer patients, higher CUL4 expression was associated with shorter overall and disease-free survival.17 Nevertheless, prognostic significance of CUL4B in GC remains unclear.
In this study, CUL4B expression at both mRNA and protein levels was higher in human GC tissues than in paired normal mucosa, in agreement with the data from Oncomine. Furthermore, we demonstrated that CUL4B expression was significantly associated with T and N classification, UICC stage and differentiation of GC, indicating that CUL4B upregulation is highly correlated with GC development and progression. Moreover, CUL4B served as a prognostic factor for DFS and OS, predicting poor survival of GC patients.
In addition, we generated CUL4B knockdown and overexpression GC cell lines, and found that CUL4B promoted cell proliferation and colony formation. Moreover, we showed that CUL4B knockdown inhibited xenograft tumor formation in nude mice, indicating that CUL4B could promote GC growth both in vitro and in vivo. Transwell assays further showed that CUL4B could facilitate GC cell invasion in vitro. It was reported that CUL4B activated Wnt/β-catenin signaling in hepatocellular carcinoma. Mechanistically, CUL4B mediated epigenetic silencing of Wnt pathway antagonists such as DKK1 and PPP2R2B by promoting the recruitment of PRC2 to their promoters to repress their expression.21 It remains unclear whether similar mechanisms may explain how CUL4B contributes to GC progression by activating Wnt/β-catenin signaling.
In conclusion, our study provided the first insight into the clinical significance of CUL4B in human GC. CUL4B expression is upregulated in GC, and elevated expression of CUL4B could promote GC cell proliferation and invasion. In addition, CUL4B is an independent predictor of unfavorable prognosis in GC, and is a potential target for GC therapy.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi:10.3322/caac.21387
2. Wu D, Zhao L, Liu Y, et al. The superiority of 256-slice spiral computed tomography angiography for preoperative evaluation of surrounding arteries in patients with gastric cancer. Onco Targets Ther. 2017;10:927–933. doi:10.2147/OTT
3. Chen PF, Wang F, Nie JY, et al. Co-expression network analysis identified CDH11 in association with progression and prognosis in gastric cancer. Onco Targets Ther. 2018;11:6425–6436. doi:10.2147/OTT.S176511
4. Sheng S, Chen Y, Li C. Outcomes of laparoscopic total gastrectomy for elderly gastric cancer patients. J Cancer. 2018;9(23):4398–4403. doi:10.7150/jca.26858
5. Li B, Zhang S, Shen H, Li C. MicroRNA-144-3p suppresses gastric cancer progression by inhibiting epithelial-to-mesenchymal transition through targeting PBX3. Biochem Biophys Res Commun. 2017;484(2):241–247. doi:10.1016/j.bbrc.2017.01.084
6. Zhang BG, Hu L, Zang MD, et al. Helicobacter pylori CagA induces tumor suppressor gene hypermethylation by upregulating DNMT1 via AKT-NFkappaB pathway in gastric cancer development. Oncotarget. 2016;7(9):9788–9800. doi:10.18632/oncotarget.7125
7. Ren J, Niu G, Wang X, Song T, Hu Z, Ke C. Overexpression of FNDC1 in gastric cancer and its prognostic significance. J Cancer. 2018;9(24):4586–4595. doi:10.7150/jca.27672
8. Van Cutsem E, Sagaert X, Topal B, Haustermans K, Prenen H. Gastric cancer. Lancet. 2016;388(10060):2654–2664. doi:10.1016/S0140-6736(16)30354-3
9. Jia S, Kobayashi R, Grewal SI. Ubiquitin ligase component Cul4 associates with Clr4 histone methyltransferase to assemble heterochromatin. Nat Cell Biol. 2005;7(10):1007–1013. doi:10.1038/ncb1300
10. Jackson S, Xiong Y. CRL4s: the CUL4-RING E3 ubiquitin ligases. Trends Biochem Sci. 2009;34(11):562–570. doi:10.1016/j.tibs.2009.07.002
11. Higa LA, Yang X, Zheng J, et al. Involvement of CUL4 ubiquitin E3 ligases in regulating CDK inhibitors Dacapo/p27Kip1 and cyclin E degradation. Cell Cycle. 2006;5(1):71–77. doi:10.4161/cc.5.1.2266
12. Lee J, Zhou P. Pathogenic role of the CRL4 ubiquitin ligase in human disease. Front Oncol. 2012;2:21. doi:10.3389/fonc.2012.00021
13. Zou Y, Liu Q, Chen B, et al. Mutation in CUL4B, which encodes a member of cullin-RING ubiquitin ligase complex, causes X-linked mental retardation. Am J Hum Genet. 2007;80(3):561–566. doi:10.1086/512489
14. Hu H, Yang Y, Ji Q, et al. CRL4B catalyzes H2AK119 monoubiquitination and coordinates with PRC2 to promote tumorigenesis. Cancer Cell. 2012;22(6):781–795. doi:10.1016/j.ccr.2012.10.024
15. Wang X, Chen Z. Knockdown of CUL4B suppresses the proliferation and invasion in non-small cell lung cancer cells. Oncol Res. 2016;24(4):271–277. doi:10.3727/096504016X14666990347473
16. Jiang T, Tang HM, Wu ZH, et al. Cullin 4B is a novel prognostic marker that correlates with colon cancer progression and pathogenesis. Med Oncol. 2013;30(2):534. doi:10.1007/s12032-013-0534-7
17. Schindl M, Gnant M, Schoppmann SF, Horvat R, Birner P. Overexpression of the human homologue for Caenorhabditis elegans cul-4 gene is associated with poor outcome in node-negative breast cancer. Anticancer Res. 2007;27(2):949–952.
18. Chen Z, Shen BL, Fu QG, et al. CUL4B promotes proliferation and inhibits apoptosis of human osteosarcoma cells. Oncol Rep. 2014;32(5):2047–2053. doi:10.3892/or.2014.3465
19. Zhang B, Zhang Y, Zhang X, Lv Y. Suspension state promotes extravasation of breast tumor cells by increasing integrin β1 expression. Biocell. 2018;42(1):17–24. doi:10.32604/biocell.2018.06115
20. Yang Y, Liu R, Qiu R, et al. CRL4B promotes tumorigenesis by coordinating with SUV39H1/HP1/DNMT3A in DNA methylation-based epigenetic silencing. Oncogene. 2015;34(1):104–118. doi:10.1038/onc.2013.522
21. Yuan J, Han B, Hu H, et al. CUL4B activates Wnt/beta-catenin signalling in hepatocellular carcinoma by repressing Wnt antagonists. J Pathol. 2015;235(5):784–795. doi:10.1002/path.4492
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.