Back to Journals » OncoTargets and Therapy » Volume 11
Upregulation of circ_0066444 promotes the proliferation, invasion, and migration of gastric cancer cells
Authors Rong D, Dong C, Fu K, Wang H, Tang W, Cao H
Received 8 November 2017
Accepted for publication 7 February 2018
Published 11 May 2018 Volume 2018:11 Pages 2753—2761
DOI https://doi.org/10.2147/OTT.S156516
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Ingrid Espinoza
Dawei Rong, Chaoxi Dong, Kai Fu, Hanjin Wang, Weiwei Tang, Hongyong Cao
Department of General Surgery, Nanjing First Hospital, Nanjing Medical University, Nanjing, People’s Republic of China
Background: Circular RNAs (circRNAs), which have closed-loop structure, are involved in the pathogenesis of human diseases including various types of carcinomas. The present study aimed to investigate the relationship between a new circular RNA named circ_0066444 and gastric cancer (GC) carcinogenesis.
Materials and methods: The circ_0066444 levels in 106 paired gastric carcinoma tissues and related adjacent normal tissues were detected by real-time quantitative reverse-transcription polymerase chain reaction. The correlation between the expression of circ_0066444 and clinicopathological features was analyzed. The impact of circ_0066444 expression on cell proliferation, invasion, as well as migration was evaluated in vitro using knockdown expression strategies. Finally, a network of circ_0066444-targeted miRNA interactions and their corresponding mRNAs was constructed.
Results: circ_0066444 was found to be significantly upregulated in 106 GC tissues as compared with paired adjacent nontumorous tissues (P=0.025), showing a high positive correlation with lymphatic metastasis (P=0.023). Furthermore, in vitro assays of the GC cell lines BGC-823 and AGS demonstrated that knockdown of circ_0066444 reduced cell proliferation, invasion, and migration significantly. Prediction and annotation revealed circ_0066444 was able to sponge to 5 miRNAs and 15 corresponding target mRNAs.
Conclusion: Our study indicated upregulation of circ_0066444 promotes gastric cell proliferation, invasion, and migration ability and might serve as a novel biomarker for GC.
Keywords: migration, invasion, proliferation, diagnoses, miRNA
Introduction
Gastric cancer (GC) is the most common digestive system tumor and remains the primary cause of human death in developing countries.1 Due to the lack of specific biomarkers, patients are diagnosed mainly in advanced stages and have a poor prognosis.2 Thus, it is necessary to identify appropriate molecular biomarkers for early-stage diagnosis and identify potential targets for GC therapy.3
Circular RNAs (circRNAs), which are different from the canonical splicing of linear RNAs, represent a novel class of conserved endogenous RNAs that are able to regulate gene expression in mammals.4,5 Some circRNAs are reported to have high tissue and cell specificity in different mammal cells.6 Growing evidence shows that circRNAs have critical functions in tumor and can have diagnostic and therapeutic potential in cancer. Recent studies have revealed that circRNAs played important roles in regulating cancer progression mainly by functioning as miRNA sponges, and these circRNA–miRNA axes are involved in various disease pathways such as cancer metastasis, apoptosis, and invasion.7,8
This study aimed to investigate the relationship between GC and a new circular RNA named circ_0066444, whose corresponding gene symbol is ADAMTS9, located at chr3 64579933-64582667 (Figure 1). The clinical significance of circ_0066444 was explored by examining the expression pattern in GC tissues and analysis of existent correlations patterns with clinicopathological features. The biological function of circ_0066444 during GC progression was further explored in in vitro assays including cell proliferation assays, cell migration assays, and invasion assays. In addition, prediction and annotation of circ_0066444 molecular function with miRNAs or mRNAs were performed on relevant databases (circinteractome and MIRDB).
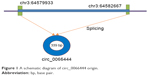  | Figure 1 A schematic diagram of circ_0066444 origin. |
Materials and methods
Patients and clinical tissue samples
A total of 106 GC tissues and corresponding adjacent nontumorous tissue samples were collected from GC patients who underwent surgical treatment between January 2013 and December 2016. Among these patients, 73 pairs of samples were from Nanjing First Hospital and the other 33 pairs were from Pukou Central Hospital, Nanjing. All tumors were staged accurately according to the tumor–node–metastasis (TNM) staging system of the International Union Against Cancer (v.8; 2016). The patients received neither radiotherapy nor chemotherapy before the operation, and their detailed characteristics including age, gender, diameter, differentiation, lymphatic metastasis, TNM stage, and common clinical biomarker levels, including CEA, CA199, CA50, and CA242, were analyzed. Written informed consent was obtained from each patient before they participated in this study. The medical ethics committee of Nanjing Medical University approved the study.
Cell line, cell culture, and transfection
Human GC cell lines MKN-45, BGC-823, MGC-803, and AGS were established from samples extracted from human GC patients by the Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences. The human gastric epithelial cell line GES-1 was obtained from the Cancer Institute and Hospital of the Chinese Academy of Medical Sciences. AGS and BGC-823 cells were transfected with 50 nM siRNAs targeting circ_0066444 (si-circ_0066444) or a scrambled negative control (si-NC) using the Lipofectamine 2000 transfection reagent (Invitrogen, Carlsbad, CA, USA) according to the instructions provided by the manufacturer. The target sequences for circ_0066444 siRNAs were as follows: siRNA-1: 5′-CCTTGGAGCTCGTGTTCCA-3′; siRNA-2 5′-TGGAGCTCGTGTTCCAGTA-3′; siRNA-3: 5′-CTCGTGTTCCAGTACCTGT-3′. After 48 h, knockdown of circ_0066444 was confirmed via quantitative real-time polymerase chain reaction (qRT-PCR).
RNA isolation, reverse transcription, and qRT-PCR
Total RNA was extracted from cancerous/noncancerous specimens stored at −80 before use or from cell lines using the RNA Isolation kit (Yifeixue, Nanjing, People’s Republic of China). RNA was reverse-transcribed into cDNAs using the Prime-Script TM one-step RT-PCR kit (Takara, Dalian, People’s Republic of China). circ_0066444 expression level was determined by qRT-PCR using the following primer sequences: forward, 5′-TTGCTCCTTGGGCGCTATAC-3′; reverse, 5′-AGAGTCCAGCGGCAAAACTA-3′. GAPDH was used as an internal control. circ_0066444 levels were normalized to GAPDH using the following primers: forward, 5′-GTCAACGGATTTGGTCTGTATT-3′; and reverse, 5′-AGTCTTCTGGGTGGCAGTGAT-3′. qRT-PCR was performed using the ABI7500 System (Applied Biosystems, Foster City, CA, USA) and the SYBR Green PCR Master Mix (Takara).
Cell proliferation assays
The MTT assay was performed according to the manufacturer’s protocol. A total of 1×104 cells were seeded in each well in 96 well plates with 90% Roswell Park Memorial Institute 1640 (RPMI 1640) and 10% fetal bovine serum. Measurements of absorbance at 570 nm were carried out using VersaMax (Molecular Devices, San Jose, CA, USA) to estimate MTT-formazan production after 24 h of incubation.
Cell migration and invasion assays
AGS and BGC-823 cells were transfected with 50 nm si-circ_0066444 or si-NC. At 24 h postinfection, infected cells were harvested and subjected to a migration assay and an invasion assay.
For migration assays, a total of 1×105 infected cells were plated in the top chamber of Transwell assay inserts (Millipore, Billerica, MA, USA) with a membrane containing pores 8 mm diameter and in 200 mL of serum-free RPMI 1640. Inserts were then placed into the bottom chamber wells of a 24-well plate containing RPMI 1640 with 10% fetal bovine serum as a chemoattractant. After 24 h of incubation, the remaining cells were removed from the top layer of the insert by scrubbing with a sterile cotton swab. Invading cells from the bottom surface were stained with 0.1% crystal violet prior to being examined, counted, and photographed using digital microscopy. Cell numbers were calculated in four random fields for each chamber, and the average value was calculated.
For invasion assays, a total of 4×104 infected cells were plated in the top chamber with a Matrigel-coated membrane. Bottom chambers were filled with conditioned medium. After a 48-h incubation period, the number of migrated cells on the lower side of the membrane was counted.
Scratch wound assays
AGS and BGC-823 cells were transfected with 50 nm si-circ_0066444 or si-NC. When cell confluence reached approximately 80% at 24 h posttransfection, wounds were created in confluent cells using a 1 mL pipette tip. Cells were then rinsed with media to remove any free-floating cells and debris. Medium was added, and culture plates were incubated at 37°C. Wound healing was observed at different time points within the scrape line, and representative scrape lines were photographed. Duplicate wells for each condition were examined, and each experiment was repeated in triplicate.
Statistical analysis
Continuous data were analyzed using an independent t-test, whereas categorical data were analyzed with the χ2 test. A receiver operating characteristic (ROC) curve was established to evaluate its diagnostic value. All statistical analyses were performed using SPSS for Windows v.17.0 (SPSS, Chicago, IL, USA). For all results, P<0.05 was considered statistically significant.
Results
Expression of circ_0066444 and clinicopathologic information in patients with GC
A total of 3 hepatocellular carcinoma tissues and their matched noncancerous tissues were collected and screened for dysregulated circRNA using human circRNA microarray reported in Xingchen Shang’s research, and the result showed circ_0066444 was significantly upregulated in hepatocellular carcinoma.9 However, its role in the occurrence and development of GC remains unknown. Therefore, we conducted a series of experiments to explore the biological functions of circ_0066444 in GC.
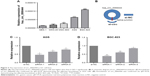
The expression levels of circ_0066444 were defined for 106 paired primary cancerous and adjacent noncancerous tissues from GC patients. A total of 88 of these cases (83%) exhibited elevated levels of circ_0066444 in tumors compared to the levels observed in adjacent nontumorous tissues (Figure 2A–D, P=0.025). No difference was observed between samples from Nanjing First Hospital and Pukou Central Hospital, Nanjing (Figure 2E). Furthermore, the ROC curve revealed that the area under the ROC curve was 0.7328 (Figure 2F) and the sensitivity and specificity of circ_0066444 were 70.75% and 68.87%, respectively.
In terms of the upregulation of circ_0066444 in GC patients, its association with clinicopathological features in GC patients was analyzed. As shown in Table 1, overexpression of circ_0066444 level was not aberrantly associated with age, sex, differentiation, or TNM stage in patients with GC but was positively associated with lymph node metastasis (P=0.023).
  | Table 1 Clinicopathological characteristics and expression of circ_0066444 |
Expression of circ_0066444 in GC cell lines and GC cell transfection
The expression of circ_0066444 in 4 GC cell lines was examined by qRT-PCR, revealing that the BGC-823 and AGS cells expressed the highest levels of circ_0066444 expression compared with GES-1 (Figure 3A). At 48 h after treatment, circ_0066444 expression was effectively knocked down in BGC-823 and AGS cells. As shown in Figure 3B–D, siRNA-1 showed the highest knockdown efficiency in GC cell line and was selected for further experiments.
Altered proliferation of GC cells by circ_0066444 expression level
Fluorouracil and mitomycin were used to detect the proliferation ability of GC BGC-823 and AGS cell lines. The result showed that knockdown of circ_0066444 inhibited cell proliferation significantly both in BGC-823 and AGS cell lines, as determined by MTT assay (Figure 4A and B).
Altered invasion and migration of GC cells by circ_0066444 expression level
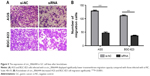
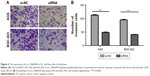
The effects of circ_0066444 on the migratory and invasive behavior of GC cell lines were assessed, demonstrating that BGC-823 and AGS cells infected with si-circ_0066444 displayed significantly lower transmembrane migration capacity compared with those infected with si-NC (Figure 5A and B). In addition, the invasion assay indicated that suppression of circ_0066444 in BGC-823 and AGS cells with si-circ_0066444 decreased invasion in the Matrigel substrate (Figure 6A and B) significantly. Furthermore, the results of a scratch wound assay demonstrated that suppression of circ_0066444 exhibited a notably lower scratch closure rate than observed in controls in BGC-823 and AGS cell lines (Figure 7A and B). These findings indicated that circ_0066444 might be closely associated with invasion and migration of GC cell lines.
circ_0066444 serves as a sponge for multiple miRNAs
To identify the miRNAs that bind to circ_0066444, we performed circRNA–miRNA interaction network prediction on circinteractome, TargetScan, and miRanda databases. A total of 5 miRNAs (ie, miR-1282, miR-1243, miR-1178, miR-638, and miR-451) and corresponding target mRNAs were predicted to have an interaction with circ_0066444 in this study. Specific details of the molecular interactions between circPVRL3 and its targets are depicted in Figure 8.
Discussion
In the present study, we first found that circ_0066444 exhibited a higher level expression in GC tissues compared with that in adjacent nontumorous tissues with a sensitivity of 70.75% and specificity of 68.87%. Furthermore, clinicopathological features of these patients showed that overexpression of circ_0066444 level was positively associated with lymph node metastasis, revealing that circ_0066444 may play a promoter role in GC. Our study was not the first circRNA which has been confirmed its value in the diagnosis of GC. Chen et al10 found circ_0000190 was downregulated in both GC tissues and plasma samples and its expression levels were significantly correlated with tumor diameter, lymphatic metastasis, distal metastasis, TNM stage, and CA19-9 levels. Apart from that, Huang11 found circ_0000745 was downregulated in GC tissues and plasma samples as well. The expression level of hsa_circ_0000745 in GC tissues correlated with tumor differentiation, while the expression level in plasma correlated with TNM stage. The fact that circRNAs are found abundantly in tissues or clinical blood samples makes circRNAs a promising diagnostic biomarker for GC screening and prognostic evaluation. However, the current methods to detect and characterize circRNAs are still limited and challengeable. Although the number of circRNAs with known functions is expanding, there are still thousands of circRNAs in GC whose functions remain unknown. A deeper understanding of circRNA biogenesis may be needed to shed light on the road of consequences of circRNAs.
Accumulating evidence indicates that circRNAs are not simply the by-products of missplicing or splicing errors, and many circRNAs have been suggested to play a role during epithelial–mesenchymal transition. For example, Zhong12 found that circRNA-MYLK might function as competing endogenous RNA for miR-29a, which could contribute to epithelial–mesenchymal transition and the development of BC through activating VEGFA/VEGFR2 and downstream Ras/ERK signaling pathway in breast cancer. Herein, we showed that inhibition of circ_0066444 expression decreased the proliferation, invasion, and migration in GC cells. Nevertheless, we could not exclude other functions of ADAMTS9 mRNA or ADAMTS9 long coding RNA because it has been implicated in cancer cells in previous studies.13,14
It has recently come to light that these circRNAs are involved in regulating gene expression by sequestering the miRNAs. For example, the expression of circLARP4 was downregulated in GC tissues. Besides, circLARP4 was mainly localized in the cytoplasm and inhibited biological behavior of GC cells by sponging miR-424.15 Therefore, we supposed that the carcinogenic mechanisms of circ_0066444 may occur through their miRNA-mediated effects on gene expression in GC as well. A total of 5 miRNAs and corresponding target mRNAs were predicted to have an interaction with circ_0066444 in this study. Taken together, we propose the hypothesis that the circRNA–miRNA–mRNA axis may be the possible mechanism promoting the development of GC, but further studies are needed to confirm this.
Limitations
Limitations should not be ignored when interpreting the results. First, all experimental samples are from two hospitals, and so it is recommended that samples from more hospital or even from patients of different ethnicities be studied. Second, experimental identification and characterization of the associated molecules, such as interactions between circRNAs and miRNAs, are suggested to be performed in future studies. Third, a previous study reported that some circRNAs can store, sort, or localize RBPs, and probably regulate the function of RBPs by acting as competing elements.6 Therefore, further studies of circ_0066444 are needed to confirm its role in this mechanism.
Conclusion
Our study indicated circ_0066444 might serve as a novel biomarker for GC and that upregulation of circ_0066444 promotes gastric cell proliferation, invasion, and migration ability. However, how circ_0066444 interacts with targeted miRNA and their corresponding mRNAs needs to be researched further. In the future, we believe that molecular mechanisms by which circRNAs function as miRNA sponges to regulate GC occurrence and development will be a promising research field.
Acknowledgments
This work was funded by grant from the Natural Science of Jiangsu Province (BK20151087) and the Development of Medical Science and Technology Foundation of Nanjing (ZKX14035) to Professor Hongyong Cao for conducting this research. Dawei Rong, Chaoxi Dong and Kai Fu are co-first authors for this study.
Disclosure
The authors report no conflicts of interest in this work.
References
Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. | ||
Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. | ||
Pan L, He M, Ma J, et al. Phase and size controllable synthesis of NaYbF4 nanocrystals in oleic acid/ionic liquid two-phase system for targeted fluorescent imaging of gastric cancer. Theranostics. 2013;3:210–222. | ||
Salzman J, Gawad C, Wang PL, et al. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7(2):e30733. | ||
Barrett SP, Salzman J. Circular RNAs: analysis, expression and potential functions. Development. 2016;143(11):1838–1847. | ||
Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. | ||
Han C, Seebacher NA, Hornicek FJ, et al. Regulation of microRNAs function by circular RNAs in human cancer. Oncotarget. 2017;8(38):64622–64637. | ||
Rong D, Tang W, Li Z, et al. Novel insights into circular RNAs in clinical application of carcinomas. Onco Targets Ther. 2017;10:2183–2188. | ||
Shang X, Li G, Liu H, et al. Comprehensive circular RNA profiling reveals that hsa_circ_0005075, a new circular RNA biomarker, is involved in hepatocellular carcinoma development. Medicine. 2016;95(22):e3811. | ||
Chen S, Li T, Zhao Q, et al. Using circular RNA hsa_circ_0000190 as a new biomarker in the diagnosis of gastric cancer. Clin Chim Acta. 2017;466:167–171. | ||
Huang M, He YR, Liang LC, et al. Circular RNA hsa_circ_0000745 may serve as a diagnostic marker for gastric cancer. World J Gastroenterol. 2017;23(34):6330–6338. | ||
Zhong Z, Huang M, Lv M, et al. Circular RNA MYLK as a competing endogenous RNA promotes bladder cancer progression through modulating VEGFA/VEGFR2 signaling pathway. Cancer Lett. 2017;403:305–317. | ||
Yao J, Zhou B, Zhang J, et al. A new tumor suppressor LncRNA ADAMTS9-AS2 is regulated by DNMT1 and inhibits migration of glioma cells. Tumour Biol. 2014;35(8):7935–7944. | ||
Ocak Z, Acar M, Gunduz E, et al. Effect of hypericin on the ADAMTS-9 and ADAMTS-8 gene expression in MCF7 breast cancer cells. Eur Rev Med Pharmacol Sci. 2013;17(9):1185–1190. | ||
Zhang J, Liu H, Hou L, et al. Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR-424-5p and regulating LATS1 expression. Mol Cancer. 2017;16(1):151. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.