Back to Journals » OncoTargets and Therapy » Volume 11
Upregulation of AKIP1 contributes to metastasis and progression and predicts poor prognosis of patients with colorectal cancer
Authors Jiang W, Yang W, Yuan L, Liu F
Received 18 September 2017
Accepted for publication 9 July 2018
Published 11 October 2018 Volume 2018:11 Pages 6795—6801
DOI https://doi.org/10.2147/OTT.S151952
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Prof. Dr. Geoffrey Pietersz
Weifang Jiang,1,* Weiji Yang,2,* Li Yuan,3 Fanlong Liu3
1Department of Surgery, The Hospital of Zhejiang University, Hangzhou 310016, China; 2Graduate Student Department, Zhejiang Chinese Medical University, Hangzhou 310053, China; 3Department of Anorectal Surgery, The First Affiliated Hospital, College of Medicine, Zhejiang University, Hangzhou 310003, China
*These authors contributed equally to this work
Background: A kinase-interacting protein 1 (AKIP1) has been reported to play an important role in the development and progression of cancer. However, the clinicopathological and biological roles of AKIP1 in colorectal cancer (CRC) remain largely unknown. The aim of this study was to investigate AKIP1 protein expression in CRC and determine the correlation between AKIP1 protein expression and clinicopathological features, as well as prognosis in CRC patients.
Materials and methods: AKIP1 protein expression was determined by immunohistochemical analysis using tissue microarrays of CRC. We also used an siRNA approach to knock down AKIP1 expression and determine the effect of AKIP1 on CRC cell migration by transwell analysis.
Results: AKIP1 expression in CRC tissue was significantly higher compared with that of noncancerous colorectal mucosa (P<0.001). Further analysis showed that AKIP1 expression was significantly associated with tumor diameter, TNM stage, and lymph node metastasis (P<0.05). Kaplan–Meier survival analysis demonstrated that patients with a positive AKIP1 expression had significantly poorer overall survival rates when compared with those with negative AKIP1 expression (P=0.031). Multivariate analysis using the Cox proportional hazard model, however, revealed that AKIP1 expression was not a significant independent prognostic factor for CRC. Transwell assay showed that the migration potential of si-AKIP1-transfected cells was significantly reduced when compared with control cells.
Conclusion: Elevated AKIP1 expression may contribute to metastasis and progression of CRC. Moreover, high AKIP1 expression in CRC significantly correlated with a patient’s shorter survival time. Therefore, AKIP1 may be a useful prognostic marker for CRC and a promising novel target for the treatment of CRC.
Keywords: AKIP1, colorectal cancer, metastasis, prognosis
Background
Colorectal cancer (CRC) is the third most common type of cancer and the second major cause of cancer-related deaths in the world.1 In recent reports, accumulated evidence has shown that CRC is a heterogeneous disease, and its molecular characteristics determine the response to treatment and, therefore, prognosis.2 The dysregulation of oncogenes or tumor suppressor genes results in changes in the biological characteristics of cancer cells, including cell growth, apoptosis, migration, invasion, and metabolism.3,4 Therefore, identifying novel biomarkers that could be applicable to the diagnosis and treatment of CRC patients is of utmost importance.
A kinase-interacting protein 1 (AKIP1), a 23 kDa protein, was originally identified in breast and prostate cancer cell lines by mRNA screens.5 Several reports have indicated that AKIP1 is localized in the cytoplasm, nucleus, and mitochondria,6 and acts as an adaptor or structural intracellular protein.7 The biochemical and biological roles of AKIP1 are not well defined. However, previous studies have revealed that AKIP1 acts as a potential factor that controls stress adaptation in the heart of mice, and that overexpression of AKIP1 in the heart protected against ischemia/reperfusion and improved cardiac function.8 In contrast, in recent studies, it was demonstrated that AKIP1 acts as an oncogene, and induced tumorigenesis and invasiveness in breast cancer, in particular, by playing a critical role in the upregulation of PKA and NF-κB signaling.9 It has been demonstrated that enhanced expression or increased activity of AKIP1 is associated with the development of human esophageal squamous cell carcinoma (ESCC), angiogenesis, and lymphangiogenesis, which indicated that AKIP1 may play a crucial oncogenic role in ESCC progression.10 Treatment results and CRC prognosis have been challenging because of liver metastasis, which has attracted increased attention. Previous studies have shown that an epithelial–mesenchymal transition (EMT) in CRC cells increases migration and invasion abilities.11 Guo et al found that AKIP1 promoted EMT via transactivating ZEB1 in non-small-cell lung cancer.12 Therefore, it is of significant interest to investigate the biological roles of AKIP1 in CRC.
In this study, we investigated AKIP1 protein expression and its correlation with clinicopathological features and clinical outcomes in patients with CRC. We envisage that our findings will provide useful information on prognosis and form a molecular basis for targeted therapies in patients with CRC.
Materials and methods
Patients and tissue samples
Tissue microarrays (TMAs) with 251 paraffin specimens of CRC were purchased from Shanghai Biochip Co., Ltd. (Shanghai, China), and were used for evaluation of AKIP1 expression by immunohistochemical analysis. The age of patients at the time of surgery ranged from 28 to 85 years (median age =66 years). TNM staging analysis following the American Joint Committee on Cancer standards identified 143 patients with stage I+II CRC and 108 patients with stage III and IV CRC. In a total of 101 patients, lymph node metastasis was observed. All patients were followed for over 5 years, and survival time was calculated from the date of surgery to the deadline for follow-up, or to the date of death. This study was approved by the ethics committee of the First Affiliated Hospital, Zhejiang University School of Medicine. All patients signed the written informed consent for this study.
Immunohistochemical analysis
TMA sections were deparaffinized using xylene followed by rehydration in graded alcohol. Antigen retrieval was performed by autoclaving the sections in citrate buffer (pH 6.0) for 3 minutes after which they were returned to room temperature and washed with PBS. Peroxidase activity was blocked by incubating the sections with 3% (v/v) H2O2 for 10 minutes at room temperature. To prevent nonspecific binding, the sections were incubated with 10% (v/v) normal goat serum for 15 minutes at room temperature and incubated overnight at 4°C with a rabbit polyclonal anti-AKIP1 antibody (1:100; antibodies-online, Aachen, Germany). After rinsing with PBS, the sections were incubated with a secondary antibody for 20 minutes at room temperature. The sections were then incubated with 3,3-diaminobenzidine. Finally, the sections were counterstained with hematoxylin, dehydrated, and mounted. As a negative control, the sections were incubated in PBS, and the primary antibody was omitted.
Immunohistochemistry scoring system
Sections were evaluated by counting the number of cells per field and were scored independently by two expert pathologists who were not aware of the clinicopathological characteristics of the CRC patients. If a score was inconsistent, the pathologists discussed and repeated the scoring to obtain an agreeable final score. The scores were grouped according to the intensity and extent of staining. The degree of positivity was based on the proportion of positively stained tumor cells: 0, no negative cells; 1, <10% positive cells; 2, 10%–50% positive cells; and 3, >50% positive cells. The intensity of staining was scored as follows: 0, negative cells; 1, weak staining; 2, moderate staining; and 3, strong staining. Consequently, the final immunohistochemical staining score was multiplied by staining intensity (0, 1, 2, 3, 4, 6, or 9). For statistical analyses, scores of 0–3 were designated negative, whereas scores of 4–9 were considered positive.
Cell culture
CRC cell lines SW480, HT-29, and HCT116 (Cell Bank of Chinese Academy of Sciences, Shanghai, China) were maintained in DMEM (Thermo Fisher Scientific, Waltham, MA, USA), supplemented with 10% FBS (HyClone, Logan, UT, USA), and incubated with 5% CO2 at 37°C.
Western blot analysis
Cell lysates from CRC cells were collected, and protein concentration was determined using the bicinchoninic acid protein assay kit (Thermo Fisher Scientific, Waltham, MA, USA). Proteins were separated by electrophoresis using an SDS-PAGE gel. Then, proteins were electro-transferred to a polyvinylidene difluoride membrane (Millipore, Bedford, MA, USA). After blocking with 5% skimmed milk for 30 minutes, the membrane was incubated with an anti-AKIP1 antibody for 2 hours at room temperature. After washing, the membrane was incubated with horseradish peroxidase-conjugated secondary antibodies. Finally, proteins were visualized using an enhanced chemiluminescence kit according to the manufacturer’s instructions.
siRNA transfection
siRNAs targeting AKIP1 and control siRNA were designed by Thermo Fisher Scientific. Cells were transfected with siRNA (final concentration was 100 nM) using Lipofectamine 3000 reagent (Thermo Fisher Scientific) as per the manufacturer’s instructions and harvested 48 hours after transfection.
RNA extraction and real-time PCR
Total RNA of cells was extracted using Trizol reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions. A SuperScriptH III First-Strand Synthesis System (Thermo Fisher Scientific) was used to synthesize cDNA. The primer sequences of AKIP1 were as follows: F, 5′-GAAGGATCCGTCGACATGGAATACTGCCTGGCGGC-3′; R, 5′-GAACTCGAGTCATACGGGGAACACCAAGTCCAC-3′. GAPDH served as an internal control. The primer sequences of GAPDH were as follows: F, 5′-GAGTCAACGGATTTGGTCGT-3′; R, 5′-TTGATTTTGGAGGGATCTCG-3′. Reactions were performed in triplicate in three separate experiments. The relative gene expression was calculated using a comparative ΔCt method.
Cell migration assay
For migration assays, cells were grown in Corning Transwell insert chambers. Briefly, cells in serum-free medium were seeded into the upper chamber. Medium containing 10% FBS was placed into the lower chamber, and this served as a chemoattractant. After incubation for 24 hours at 37°C, the non-migrated cells attached to the upper surface of the membrane were softly wiped by a cotton swab, whereas the cells that migrated through the membrane were washed and incubated with 0.1% crystal violet for 20 minutes at room temperature. Migrated cells in three random fields were counted at ×200 magnification.
Statistical analysis
Statistical analyses were performed using SPSS version 13.0 software (SPSS, Chicago, IL, USA) Chi-square tests (Pearson as appropriate) or Fisher’s exact probabilities were used to evaluate the correlation between AKIP1 expression and the baseline clinical characteristics of patients with CRC. Kaplan–Meier survival curve was generated by plotting patient survival probability and was analyzed by the log-rank test. A Cox regression model was used for the analysis of independent prognostic risk factors. Paired t-test was used to analyze the effect of AKIP1 siRNA on CRC cell lines. Statistical tests were two-sided, and P<0.05 was considered statistically significant.
Results
Expression of AKIP1 in CRC and noncancerous colorectal mucosa
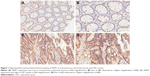
Immunohistochemical analysis was confined to the cytoplasm of cells. Typical diffuse cytoplasmic staining of the AKIP1 protein is shown in Figure 1. Evaluation of the staining by two independent pathologists indicated that 15 cases of CRC were discordant, and the kappa value was 0.928. This suggested that the interobserver reproducibility of the evaluation was sufficient. Expression of the AKIP1 protein was detected in 55.4% (139/251) of CRC-derived tissue and in 4.8% (12/251) of noncancerous colorectal mucosa. Moreover, the AKIP1 expression in CRC-derived tissue was significantly higher compared with that in noncancerous colorectal mucosa (P<0.001).
Relationship between AKIP1 level and clinicopathological features of CRC
In this study, the association between AKIP1 protein expression and the clinicopathological features was evaluated. As summarized in Table 1, AKIP1 expression was significantly associated with TNM stage and lymph node metastasis. CRC patients with a high TNM stage (stages III+IV) showed significantly higher expression of AKIP1 compared with those with a low TNM stage (stages I+II) (Table 1). The Spearman correlation coefficient of AKIP1 expression with TNM stage and lymph node metastasis was 0.294 and 0.279 (P<0.05), respectively. No significant association was observed between AKIP1 expression and gender, age, differentiation, distant metastasis, and liver metastasis.
  | Table 1 Relationship between AKIP1 expression and clinicopathological features of colorectal cancer |
Correlation of AKIP1 expression with overall survival
The 3- and 5-year cumulative survival rates were 63.6% and 54.5% for patients with positive AKIP1 expression, and were 66.6% and 63.4% for those with negative AKIP1 expression. The mean survival time for patients with positive expression of AKIP1 was 59.03±2.94 months, and for those with negative expression of AKIP1, 65.05±3.16 months.
Kaplan–Meier survival analysis and log-rank test demonstrated that patients with positive expression of AKIP1 in their tumors had a significantly poorer overall chance of survival when compared with those in whom the tumor did not express AKIP1 (P=0.031, log-rank test; Figure 2).
Multivariate analysis of prognosis using Cox proportional hazard model
The results of univariate analysis showed that differentiation (P=0.024), TNM stage (P<0.001), distant metastasis (P<0.001), liver metastasis (P<0.001), and AKIP1 expression (P=0.031) were significantly associated with the overall survival rate in patients with CRC (Table 2). Thus, these data indicated that AKIP1 may be a promising prognostic factor for CRC.
  | Table 2 Univariate analysis of the correlation between clinicopathological parameters and survival of patients with colorectal cancer |
To evaluate whether AKIP1 expression could provide prognostic information independent of clinical variables, multivariable Cox proportional hazard analysis was performed for all variables examined in the univariate analysis (Table 3). We determined that TNM stage (HR =3.237; 95% CI 1.155–9.074; P=0.025), liver metastasis (HR =5.448; 95% CI 1.019–29.110; P=0.047), and differentiation (HR =1.396; 95% CI 1.014–1.922; P=0.041) were significantly independent prognostic factors for survival in CRC, whereas AKIP1 expression was not (P=0.314).
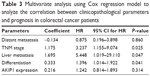  | Table 3 Multivariate analysis using Cox regression model to analyze the correlation between clinicopathological parameters and prognosis in colorectal cancer patients |
Downregulation of AKIP1 inhibits the migration of CRC cells
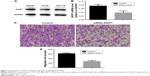
In this study, we determined AKIP1 protein levels in CRC cell lines and showed that CRC cells expressed high levels of AKIP1 (Figure 3A). To evaluate the impact of AKIP1 expression on the migration abilities of CRC cells, the expression of AKIP1 was knocked down using a specific siRNA in SW480 CRC cells, which showed a relatively high expression of AKIP1. Silencing of AKIP1 expression was verified by quantitative reverse transcription PCR (P=0.001; Figure 3B). The migration potential of si-AKIP1-transfected cells was significantly reduced compared with the respective control cells (P=0.008; Figure 3C and D).
Discussion
AKIP1 was cloned from a subtractive hybridization library that was derived from matched breast tumor and normal breast cell mRNA.5,6 In recent years, the role of AKIP1 in carcinogenesis and tumor development has become a hot topic. AKIP1 was reported to modulate NF-κB and PKA activities,6,13 which resulted in the induction of apoptosis and the reduction of tumor progression.5,14–16 In contrast, several studies revealed that AKIP1 stimulated neovascularization and tumor growth. For example, Lin et al10 demonstrated that Nkx2-8 repressed NF-κB activity by restraining nuclear localization of NF-κB p65 via downregulation of AKIP1 in ESCC cells. In their study, they also showed that overexpression of AKIP1 induced ESCC angiogenesis and lymphangiogenesis. Furthermore, previous studies have demonstrated that high AKIP1 levels could transcriptionally upregulate VEGF-C by interaction of AKIP1 with its promoter through cooperation with multiple transcriptional factors.10 This apparent discrepancy suggested that the role of AKIP1 may vary in different cell types and clinical situations, or that in a normal physiological setting, the protein may exhibit feedback regulatory properties to modulate cell signaling. Overexpressed AKIP1, which can activate NF-κB, was highly observed in breast and prostate cancers,5 suggesting that upregulation of AKIP1 may contribute to the development and progression of cancer. However, the clinical significance and biological role of AKIP1 in cancer progression remain unclear.
In this study, we evaluated the expression of AKIP1 and its prognostic implications in CRC patients. When compared with adjacent noncancerous colorectal mucosa, the expression of AKIP1 was upregulated in CRC tissue, and elevated AKIP1 expression was significantly associated with the TNM stage, thereby suggesting that AKIP1 overexpression may contribute to the progression of CRC. In a previous study, the oncogenic role of AKIP1 in tumor development was demonstrated.10,17 Leung and Ngan showed that AKIP1 was a protein partner of TAp73, and the two cooperated with each other to sensitize the response of cervical cancer cells to radiotherapy.15 Moreover, transwell assays indicated that the migration potential of si-AKIP1-transfected cells was significantly reduced. Thus, our data are consistent with the findings published in previous reports, which suggested that AKIP1 may play a critical role in CRC cell proliferation and migration.
Mo et al17 revealed that AKIP1 overexpression significantly correlated with poor overall survival and recurrence-free survival in breast cancer patients. In a study by Lin et al, it was demonstrated that AKIP1 expression significantly correlated with ESCC progression and a shorter survival time in patients.10 In our present study, we performed Kaplan–Meier survival analysis to demonstrate that patients with negative AKIP1 expression had a significantly better overall survival compared with those with positive AKIP1 expression. However, multivariable Cox proportional hazard analysis revealed that AKIP1 expression was not an independent prognostic factor for survival in CRC patients. Thus, these results indicated that AKIP1 may represent a prospective prognostic indicator of CRC. Because of the limitations of the scoring method and sample collection, additional analysis is required to verify the role of AKIP1, as well as its mechanisms underlying tumorigenesis and progression of CRC.
Conclusion
In this study, we demonstrated that elevated expression of AKIP1 may contribute to metastasis and progression of CRC, findings heightened by the positive correlation between high levels of AKIP1 expression and a shorter survival time in CRC patients. Therefore, AKIP1 may be a promising prognostic marker for CRC and a novel target in the treatment of this disease.
Acknowledgment
This work was supported by the grant from the National Natural Science Foundation of China (No 81372463).
Disclosure
The authors report no conflicts of interest in this work.
References
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. | ||
Sridharan M, Hubbard JM, Grothey A. Colorectal cancer: how emerging molecular understanding affects treatment decisions. Oncology. 2014;28(2):110–118. | ||
Ordóñez-Morán P, Dafflon C, Imajo M, Nishida E, Huelsken J. HOXA5 Counteracts Stem Cell Traits by Inhibiting Wnt Signaling in Colorectal Cancer. Cancer Cell. 2015;28(6):815–829. | ||
Liu W, Li H, Hong SH, Piszczek GP, Chen W, Rodgers GP. Olfactomedin 4 deletion induces colon adenocarcinoma in ApcMin/+ mice. Oncogene. 2016;35(40):5237–5247. | ||
Kitching R, Li H, Wong MJ, Kanaganayakam S, Kahn H, Seth A. Characterization of a novel human breast cancer associated gene (BCA3) encoding an alternatively spliced proline-rich protein. Biochim Biophys Acta. 2003;1625(1):116–121. | ||
Sastri M, Barraclough DM, Carmichael PT, Taylor SS. A-kinase-interacting protein localizes protein kinase A in the nucleus. Proc Natl Acad Sci U S A. 2005;102(2):349–354. | ||
León DA, Cànaves JM. In silico study of breast cancer associated gene 3 using LION Target Engine and other tools. Biotechniques. 2003;35(6):1222–1226, 1228, 1230–1231. | ||
Sastri M, Haushalter KJ, Panneerselvam M, et al. A kinase interacting protein (AKIP1) is a key regulator of cardiac stress. Proc Natl Acad Sci U S A. 2013;110(5):E387–E396. | ||
Gao N, Hibi Y, Cueno M, Asamitsu K, Okamoto T. A-kinase-interacting protein 1 (AKIP1) acts as a molecular determinant of PKA in NF-kappaB signaling. J Biol Chem. 2010;285(36):28097–28104. | ||
Lin C, Song L, Liu A, et al. Overexpression of AKIP1 promotes angiogenesis and lymphangiogenesis in human esophageal squamous cell carcinoma. Oncogene. 2015;34(3):384–393. | ||
Shen H, Ma JL, Zhang Y, et al. Integrin-linked kinase overexpression promotes epithelial-mesenchymal transition via nuclear factor-κB signaling in colorectal cancer cells. World J Gastroenterol. 2016;22(15):3969–3977. | ||
Guo X, Zhao L, Cheng D, Mu Q, Kuang H, Feng K. AKIP1 promoted epithelial-mesenchymal transition of non-small-cell lung cancer via transactivating ZEB1. Am J Cancer Res. 2017;7(11):2234–2244. | ||
Gao N, Asamitsu K, Hibi Y, Ueno T, Okamoto T. AKIP1 enhances NF-kappaB-dependent gene expression by promoting the nuclear retention and phosphorylation of p65. J Biol Chem. 2008;283(12):7834–7843. | ||
Zimmerman R, Peng DJ, Lanz H, et al. PP2A inactivation is a crucial step in triggering apoptin-induced tumor-selective cell killing. Cell Death Dis. 2012;3:e291. | ||
Leung TH, Ngan HY. Interaction of TAp73 and breast cancer-associated gene 3 enhances the sensitivity of cervical cancer cells in response to irradiation-induced apoptosis. Cancer Res. 2010;70(16):6486–6496. | ||
Gao F, Cheng J, Shi T, Yeh ET. Neddylation of a breast cancer-associated protein recruits a class III histone deacetylase that represses NFkappaB-dependent transcription. Nat Cell Biol. 2006;8(10):1171–1177. | ||
Mo D, Li X, Li C, et al. Overexpression of AKIP1 predicts poor prognosis of patients with breast carcinoma and promotes cancer metastasis through Akt/GSK-3β/Snail pathway. Am J Transl Res. 2016;8(11):4951–4959. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.