Back to Journals » OncoTargets and Therapy » Volume 14
Upregulated MicroRNA-483-3p is an Early Event in Pancreatic Ductal Adenocarcinoma (PDAC) and as a Powerful Liquid Biopsy Biomarker in PDAC
Authors Shao H , Zhang Y, Yan J, Ban X, Fan X, Chang X, Lu Z, Wu Y, Zong L, Mo S, Yu S, Chen J
Received 27 October 2020
Accepted for publication 30 January 2021
Published 25 March 2021 Volume 2021:14 Pages 2163—2175
DOI https://doi.org/10.2147/OTT.S288936
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Takuya Aoki
Huilin Shao, Yue Zhang, Jie Yan, Xinchao Ban, Xiaojie Fan, Xiaoyan Chang, Zhaohui Lu, Yan Wu, Liju Zong, Shengwei Mo, Shuangni Yu, Jie Chen
Department of Pathology, Peking Union Medical College Hospital, Chinese Academy of Medical Sciences, Peking Union Medical College, Beijing, People’s Republic of China
Correspondence: Shuangni Yu; Jie Chen Tel +86 10 69159375
; +86 10 69159389
Email [email protected]; [email protected]
Background: There is an urgent need for the development of effective noninvasive biomarkers for early pancreatic cancer diagnosis. MicroRNAs (miRNAs) are promising candidates that can be identified in peripheral blood and can act as “liquid biopsy” biomarkers. miR-483-3p is overexpressed in the tumor tissue of pancreatic duct adenocarcinoma, but its potential as noninvasive biomarker remains unknown.
Methods: We conducted locked nucleic acid in situ hybridization (LNA-ISH) for miR-483-3p in archival tissues of 107 patients with PDAC. We also used immunohistochemistry to evaluate SMAD4 expression, the putative miR-483-3p target gene. miR-483-3p expression level was also assessed using quantitative real-time PCR (qRT-PCR) in serum and serum exosome samples from 63 patients with PDAC and 22 healthy individuals.
Results: LNA-ISH showed that miR-483-3p was overexpressed in PDAC and PanIN tissues compared to normal pancreatic duct cells. miR-483-3p expression levels correlated with increases in PanIN lesion grade. miR-483-3p expression negatively correlated with Smad4 expression (γ=− 0.770, p< 0.0001) in PDAC and PanIN tissues. Circulating miR-483-3p levels were significantly elevated in the serum and serum exosomes of PDAC patients compared to healthy controls (p< 0.0001 and p< 0.01, respectively). Specifically, serum miR-483-3p levels were able to distinguish patients with early stage (≤ 2cm) PDAC from healthy controls with an AUC of 0.83 [95% CI, 0.70– 0.96]. Higher serum exosomal miR-483-3p levels predicted worse survival in PDAC patients and serum exosomal miR-483-3p also proved to be an independent prognostic factor for PDAC (hazard ratio = 3.307; 95% CI=1.104 to 9.903; p=0.033). In vitro studies also showed that miR-483-3p promoted pancreatic cancer cell migration and invasion.
Conclusion: miR-483-3p overexpression occurs early in PDAC development and is present in premalignant PanIN lesions. Serum miR-483-3p may act as an early PDAC diagnostic biomarker and serum exosomal miR-483-3p may be a PDAC prognostic biomarker.
Keywords: pancreatic ductal adenocarcinoma, exosome, microRNA-483-3p, liquid biopsy, biomarker
Introduction
Pancreatic cancer is an extremely lethal malignancy and the seventh leading cause of cancer-related death worldwide.1 Pancreatic cancer morbidity increases each year and the five-year overall survival rate is only 9% in 2019 in the United States, the worst of all cancer types.2 This poor prognosis is often attributed to late diagnosis, as most of the patients present with advanced disease that is unresectable.3 Because early detection can drastically improve patient outcomes, there is an urgent need to develop sensitive and specific biomarkers for early detection.
Pancreatic ductal adenocarcinoma (PDAC) accounts for more than 90% of pancreatic cancers and is believed to arise from noninvasive precursor lesions.4 Pancreatic intraepithelial neoplasia (PanIN) is the most common noninvasive precursor to PDAC.5 PanINs are microscopic epithelial neoplasms in the small pancreatic ducts that exhibit a varying cytological and architectural atypia. PanINs are classified into three grades based on the degree of dysplasia: PanIN-1, PanIN-2 and PanIN-3.6,7 PanINs represent a curable stage prior to PDAC development and are the important targets for early detection.
microRNAs (miRNAs) are short, single strand non-coding RNAs that are frequently dysregulated in a variety of human cancers.8–10 miRNAs regulate post-transcriptional gene expression primarily by binding to the 3ʹ untranslated region of target messenger RNAs (mRNAs).11 miRNAs have been implicated as both oncogenes and tumor suppressor genes in cancer12 and are promising non-invasive biomarker candidates. Our team has reported that miR-483-3p expression is elevated in tumor tissues of PDAC and a small subset of PanIN lesions, and is associated with PDAC prognosis.13 Up-regulation of miR-483-3p has been reported in colorectal, breast, and pancreatic cancers.14 SMAD4 has been identified as a target gene of miR-483-3p in pancreatic cancer15 and miR-483-3p has been shown to promote tumor cell proliferation in pancreatic cancer in preclinical models.13,15
Exosomes are 40–100 nm sized extracellular membrane vesicles that carry bioactive substances such as lipids, proteins and nucleic acids (ie, mRNA and miRNA).16,17 Nearly all mammalian cells secrete exosomes, including cancer cells, which mediate cell-to-cell communication.18,19 miRNAs are thought to be the pivotal functional molecules carried in exosomes.20 Increasing evidence suggests that exosomes play important roles in cancer biology by carrying miRNA that affects tumor proliferation, angiogenesis, energy metabolism, pre-metastatic niche formation and dissemination.21–26 Other recent studies have also suggested the potential value of exosomal miRNAs as novel cancer biomarkers in liquid biopsies.27–29
In this study, we evaluated the potential for circulating miR-483-3p to act as PDAC biomarkers that can be identified via liquid biopsy. We quantified circulating miR-483-3p expression levels in serum and serum exosome samples from PDAC patients and healthy controls using TaqMan microRNA assays. Additionally, we analyzed miR-483-3p expression in matched PDAC and PanIN lesion tissue specimens from patients with PDAC. We demonstrated that miR-483-3p is overexpressed in PanIN and PDAC tissues and is significantly elevated in both serum and serum exosomes of PDAC patients. Our findings suggest that serum miR-483-3p levels could serve as a potential liquid biopsy biomarker for early detection of PDAC. We also found that exosomal miR-483-3p levels in serum may be a useful PDAC prognostic biomarker.
Materials and Methods
Patients, Tissue Samples, and Serum Samples
Patients, tissue samples, and serum samples were conducted in accordance with the Declaration of Helsinki. The study was approved by the Medical Ethics Committee of the Peking Union Medical College Hospital (PUMCH; Beijing, China). A total of 107 consecutive formalin-fixed, paraffin-embedded PDAC specimens were retrieved from the archival files of the Department of Pathology at PUMCH (Table S1). All cases were reviewed independently by two pathologists to confirm a diagnosis of PDAC. Multiple hematoxylin-eosin-stained slides of each case were screened microscopically for PanIN lesions adjacent to PDAC lesions. The PanIN lesions were classified according to the WHO 3-tiered grading system.6 In total, we identified 272 PanIN lesions, which included 117 PanIN-1, 97 PanIN-2, and 58 PanIN-3. Matched adjacent non-pathologic pancreas was available for all cases. The 107 PDAC patients from whom tissue was collected underwent surgical resection prior to initial non-surgical treatment at the PUMCH during a time period from 2013 to 2017. Clinical data were collected and all patients received follow-up care (follow-up time 6 to 65 months). Overall survival time was calculated from the surgical resection date to the death date or the date of last follow-up. Tumor staging was assessed using the American Joint Committee on Cancer (AJCC) 8th edition staging system for pancreatic cancer.30 Unstained 5 μm sections were cut from paraffin blocks for additional analysis.
Preoperative serum samples were collected from 63 PDAC patients (from the cohort of 107 patients mentioned above), as well as 22 healthy controls in 2017 (Table S5). All serum samples were stored at −80°C until time of analysis. All study participants provided informed consent.
In situ Hybridization
LNA-ISH was conducted on archival PDAC and PanIN lesions with LNATM probes against miR-483-3p (Exiqon) according to a previously described protocol.13 A U6 probe and scrambled LNA probe were used as positive and negative controls, respectively. All probes were labeled with digoxigenin at both the 5ʹ and 3ʹ ends. Briefly, the slides were deparaffinized, digested with proteinase K, fixed in 4% paraformaldehyde, and treated with 0.1 M triethanolamine. After rinsing in PBS, the slides were pre-hybridized without a probe for two hours at 56°C, and subsequently incubated in hybridization buffer which contained the digoxigenin-labeled LNATM probe at 56°C for 16 h. After stringent washing with 0.2 × SSC at the hybridization temperature, the slides were incubated with blocking buffer for one hour and reacted with anti-digoxigenin-AP Fab fragments (1:1000; Roche) overnight at 4°C. The reaction products were visualized using NBT/BCIP Ready-to-Use tablets (Roche) in the dark for 24 h. Slides were then stained with 0.1% nuclear fast red and mounted using mounting medium. Results were analyzed using light microscopy. The ISH results were scored as 0 (no staining, negative), 1 [weak or focal staining (ie, <50% cells), positive], or 2 [strong and diffuse staining (ie, ≥50% of cells), positive].13
Immunohistochemical Staining
Unstained tissue sections were deparaffinized, then boiled in 0.01 M citric acid buffer (pH 6.0) under high pressure for 10 min. Slides were then treated with 3% hydrogen peroxide for 10 min to inactivate endogenous peroxidase. After incubating with anti-Smad4 antibody (1:150; ab40759, Abcam) at 4°C overnight, the slides were washed and labeled with HRP-conjugated anti-rabbit secondary antibody for one hour at room temperature. Staining was then visualized using 3, 3ʹ-diaminobenzidine. The anti-Smad4 antibody was replaced with rabbit serum as negative control.
The immunostaining of PanIN and PDAC lesions was evaluated independently by two pathologists and a consensus agreement was obtained for all lesions. Slides were scored as 0 (negative), 1 (focally positive), or 2 (diffusely positive) according to the criteria used in a previous study.31 Normal pancreatic ducts and acini served as positive internal controls for each slide.
Cell Lines
The human pancreatic cancer cell lines AsPC1 and PANC1 were recently purchased from the ATCC and not revalidated for this work. Culture conditions included DMEM medium (Corning, USA) supplemented with 10% fetal bovine serum.
Exosome Isolation
We extracted exosomes from 200 μL of patient serum using ExoQuick™ Serum Exosome Precipitation Solution (System Biosciences) per the manufacturer’s instructions. Briefly, serum was thawed on ice and centrifuged at 3000 g for 15 min to remove any cells or cellular debris. 50 μL ExoQuick solution was added into the 200 μL serum sample and mixed thoroughly. The mixture was kept undisturbed at 4°C overnight and then centrifuged at 1500 g for 30 minutes the next day. The supernatant was then discarded and exosomes were suspended in 100 μL PBS (Corning, USA) for analysis.
miRNA Extraction from Serum and Exosomes
Total RNA (including miRNA) was extracted from 200 μL of serum or exosomes (isolated from 200 μL serum) using the MagMAX™ mirVana™ Total RNA Isolation Kit (Thermo Fisher Scientific, Lithuania), per the manufacturer’s protocol. Twenty-five fmol synthetic miRNA spike-in control (cel-miR-39, RiboBio) was added to each sample after denaturation to normalize variation between samples that might be generated during the miRNA extraction and subsequent quantitative reaction.32 The extracted RNA was examined using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific) to assess the purity and concentration of the RNA.
Quantitative Real-Time PCR
miRNA levels in serum and serum exosome samples were evaluated via quantitative real-time PCR (qRT-PCR) using the TaqMan MicroRNA Assay (Thermo Fisher Scientific). First, total RNA was reverse transcribed into cDNA using a TaqMan™ MicroRNA Reverse Transcription Kit (Thermo Fisher Scientific), per the manufacturer’s instructions. Quantitative PCR was then performed on an ABI 7500 Real-time PCR System (Applied Biosystems, CA) following these cycling conditions: 50°C for 2 minutes (min), then 95°C for 10 min, followed by 40 cycles of 95°C for 15 seconds (sec) and 60°C for 1 min. The relative expression level of miR-483-3p was determined using the 2−ΔΔCt method.33 The miRNA spike-in (cel-miR-39) was used as a normalizing control. Each sample was assayed in triplicate.
In vitro Cell Migration and Invasion Assays
Cells were transfected with an hsa-miR-483-3p mimic or inhibitor with negative controls (NC) (RioboBio) via a Lipofectamine 2000 (Invitrogen) reagent, per the manufacturer’s instructions. Cell migration and invasion assays were conducted using 6.5 mm chambers with 8.0 µm pores either coated or uncoated with Matrigel (Corning). Twenty-four hours after transfection, cells were added to the upper chamber and 600 μL growth medium was added to the lower chamber. Cells were incubated for 24 hours, then fixed with 4% PFA and stained with 0.1% crystal violet. Cells that migrated to the lower chamber of the membrane were counted.
Statistical Analysis
The frequency of miR-483-3p or Smad4 expression in PDAC and PanIN lesions was compared using Pearson’s chi-square tests or Fisher’s exact tests. P values were adjusted for multiple comparisons. The Mann–Whitney U-test was performed to compare the serum and exosomal miR-483-3p levels between two groups. PDAC patients were divided into “low level” and “high level” groups based on their median serum or exosomal miR-483-3p expression level. Correlations between clinicopathologic parameters and serum/exosomal miR-483-3p levels were analyzed using Pearson’s chi-square tests or Fisher’s exact tests. Receiver operating characteristic (ROC) curves were generated and the area under the ROC curve (AUC) was calculated to evaluate the diagnostic potential of serum/exosomal miR-483-3p levels. Kaplan–Meier analyses and Log rank tests were performed to compare overall patient survival. Cox proportional hazards regression analysis was used to assess the association between multiple risk factors and survival time and identify prognostic prediction factors. Statistical analyses were conducted with SPSS Statistics 21.0, GraphPad Prism 7.0, and Excel. All tests were two-tailed and P<0.05 was considered statistically significant.
Results
miR-483-3p is Overexpressed in Precancerous PanIN and PDAC Lesions
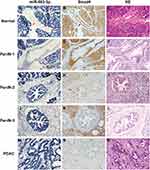
We analyzed miR-483-3p expression using LNA-ISH in formalin-fixed, paraffin-embedded tissue sections of PanIN and PDAC lesions from 107 patients with PDAC. In total, we identified 272 PanIN lesions of all histological grades among the 107 PDAC cases. miR-483-3p was expressed in 64.1% (75 of 117) of PanIN-1, 84.5% (82 of 97) of PanIN-2, and 96.6% (56 of 58) of PanIN-3 lesions, and 100% (107 of 107) of PDAC lesions. However, it was not expressed in normal pancreatic ducts (Table 1, Figure 1).
 |
Table 1 Frequency of miR-483-3p Expression in PanIN Lesions and PDAC by LNA-ISH |
The percentage of miR-483-3p positive lesions was significantly higher in PanIN-2, PanIN-3, and PDAC lesions compared to PanIN-1 lesions (two-tailed χ2 test, p=0.001, p<0.0001, and p<0.0001, respectively) (Table S2). The percentage of miR-483-3p positive lesions was also significantly higher in PDAC lesions than in PanIN-2 (two-tailed χ2 test, p<0.0001) lesions. In 32 of the 107 cases, a prominent, graded increase in miR-483-3p staining intensity was observed that corresponded with histological change from normal duct to PanINs to PDAC. LNA-ISH results showed that aberrant expression of miR-483-3p is associated with the development of PDAC. miR-483-3p expression was up-regulated as early as the PanIN-1 stage and showed a progressive increase of the expression frequency in PanIN-2/3 lesions and PDAC (Figure 1A, C, E, G and I). Overexpression of miR-483-3p was observed in both epithelial cells and adjacent stromal fibroblasts.
Expression of miR-483-3p Negatively Correlated with That of Smad4 in PDAC and PanIN Lesions
Prior studies have shown that SMAD4 is a target gene of miR-483-3p in pancreatic cancer and other cell types.15,34,35 We performed immunohistochemical staining of Smad4 on serial sections from the aforementioned archival tissues. In all 107 cases, normal pancreatic tissue including pancreatic acini, ducts, and islets of Langerhans showed a strong cytoplasmic staining pattern for Smad4. Smad4 was expressed in 99.1% (111 of 112) of PanIN-1, 97.8% (87 of 89) of PanIN-2, and 73.7% (42 of 57) of PanIN-3 lesions (Table S3, Figure 1). In contrast, 53.3% (57 of 107) of PDAC lesions completely lost Smad4 expression. The percentage of Smad4 positive lesions was significantly higher in PanIN-1/2 lesions compared to PanIN-3 and PDAC lesions. (PanIN-1 vs PanIN-3, p<0.0001; PanIN-1 vs PDAC, p<0.0001; PanIN-2 vs PanIN-3, p<0.0001; PanIN-2 vs PDAC, p<0.0001; PanIN-3 vs PDAC, p=0.001) (Table S4, Figure 1B, D, F, H and J).
miR-483-3p and Smad4 expression were mutually exclusive in PDAC tissues and the adjacent PanIN lesions, as well as normal pancreatic ducts. In the 107 PDAC patients, miR-483-3p and Smad4 protein expression was negatively correlated in both PanIN and PDAC lesions (γ=−0.770, p<0.0001) (Table S6), which supports the conclusion that miR-483-3p expression may inhibit Smad4 during the development of PDAC.
miR-483-3p Promotes Pancreatic Cancer Cell Migration and Invasion
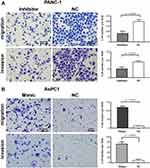
miR-483-3p has been reported to promote pancreatic cancer cell proliferation and colony formation in vitro.13,15 We further examined the effects of miR-483-3p on pancreatic cancer cell migration and invasion. We transfected the pancreatic cancer cell line Panc-1 with a miR-483-3p inhibitor and control miRNA. We also transfected the pancreatic cancer cell line AsPC1 with a miR-483-3p mimic and negative control miRNA. In Panc-1 cells, the miR-483-3p inhibitor significantly suppressed cell migration and invasion in in vitro transwell assays, relative to the negative control miRNA (p<0.01, p<0.01; respectively) (Figure 4A). In AsPC1 cells, a miR-483-3p mimic significantly increased cell migration and invasion in similar assays, compared to a negative control miRNA (p<0.01, p<0.01; respectively) (Figure 4B).
Circulating miR-483-3p Levels are Elevated in PDAC Patients and Can Distinguish PDAC Patients from Healthy Subjects
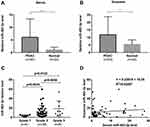
Next, we examined the expression of circulating miR-483-3p in serum samples of 63 PDAC patients and 22 healthy controls. miR-483-3p level was evaluated in exosomes and serum using RT-qPCR. Serum and exosomal miR-483-3p levels were significantly higher in PDAC patients as compared to healthy subjects (p<0.0001 and p<0.01, respectively) (Figure 2A and B). Interestingly, serum and exosomal miR-483-3p levels were not correlated in PDAC patients (Figure 2D).
We then assessed the diagnostic potential of miR-483-3p in PDAC using ROC curves. Serum miR-483-3p levels could differentiate PDAC patients from healthy subjects with a ROC curve area (AUC) of 0.81 [95% CI, 0.71–0.91] (Figure 3A). The sensitivity and specificity were 74.6%, and 77.3%, respectively, with a cut-off value of 1.2839. Importantly, serum miR-483-3p could also distinguish PDAC patients with smaller tumors (≤2cm) from healthy controls with an AUC of 0.83 [95% CI, 0.70–0.96] and the sensitivity and specificity were 85.7%, and 72.7%, respectively, with a cut-off value of 1.0722. Serum miR-483-3p had an AUC of 0.79 [95% CI, 0.65–0.93] when differentiating Stage I PDAC from healthy controls (Figure 3A). With a cut-off value of 1.0722, the sensitivity and specificity were 72.2%, and 72.7%, respectively. Exosomal miR-483-3p had an AUC of 0.69 [95% CI, 0.58–0.81] (Figure 3A) when differentiating PDAC patients from healthy controls, indicating that the diagnostic value of serum miR-483-3p level was greater than that of exosomal miR-483-3p levels. However, combining serum miR-483-3p levels with exosomal miR-483-3p levels slightly improved the diagnostic capability with an AUC of 0.84 [95% CI, 0.76–0.93] (Figure 3A), compared to serum miR-483-3p levels alone, the combination did not improve the sensitivity or specificity to identify PDAC patients.
We compared the diagnostic performance of serum miR-483-3p with CA19-9, a PDAC serum marker widely used in clinic. In ROC curve analysis, CA 19–9 generated an AUC value of 0.87 [95% CI, 0.79–0.94] to discriminate PDAC from healthy controls; and the sensitivity and specificity were 76.2%, and 100%, respectively. The AUC values of serum miRNA-483-3p and the combination of serum and exosomal miRNA-483-3p were slightly lower than that of CA 19–9. Combined CA 19–9 and serum miRNA-483-3p levels yielded an AUC value of 0.94 [95% CI, 0.89–0.99], with 82.5% sensitivity and 95.5% specificity, which showed good performance to identify PDAC.
Circulating miR-483-3p Levels are Associated with Clinicopathological Characteristics and Can Predict Survival of PDAC Patients
We then performed a correlation analysis to understand the relationship between circulating miR-483-3p levels and clinicopathological findings in PDAC. Sixty-three PDAC patients were divided into two groups on the basis of their median serum or exosomal miR-483-3p level. We found that high exosomal miR-483-3p level was significantly associated with poorer tumor differentiation, higher N stage, and higher overall stage (p=0.023, p=0.019, and p=0.022, respectively) (Figure 2C and Table 2). Patients with poorly differentiated PDAC had higher exosomal miR-483-3p levels versus patients with well-differentiated PDAC (p=0.0122). However, no significant associations were found between tumor size, T stage, age, gender, and exosomal miR-483-3p expression (Table 2). Serum miR-483-3p levels also did not significantly correlate with any clinicopathologic characteristics.
 |
Table 2 Correlation Between Circulating miR-483-3p Expression and Clinicopathologic Characteristics of PDAC Patients |
To determine whether circulating miR-483-3p levels could predict patient outcomes after pancreatic resection, we performed Kaplan–Meier survival analysis. Overall survival (OS) was significantly worse in patients with high exosomal miR-483-3p levels (at or above the median) compared to patients with low levels (below the median) (follow-up time 6 to 65 months, p=0.0064; Log rank test) (Figure 3B). In contrast, serum miR-483-3p levels showed no association with patient outcome (Figure 3B). Univariate Cox regression analysis revealed that worse survival outcomes were significantly associated with tumor size (>2.0cm), overall stage (>I), and high exosomal miR-483-3p level. Furthermore, multivariate Cox analysis demonstrated that high exosomal miR-483-3p level was an independent prognostic indicator for PDAC patients (HR=3.307; 95% CI=1.104 to 9.903; p=0.033) (Table 3).
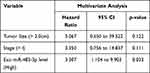 |
Table 3 Multivariate Analysis of Circulating miR-483-3p Levels and Other Clinicopathologic Factors in PDAC Patients |
Discussion
The survival rate of patients with PDAC has shown minimal improvement over the last three decades.36 An important limiting factor is that most patients are asymptomatic or have non-specific symptoms in early stages of PDAC development. Identifying molecules involved in the early stages of pancreatic cancer pathogenesis can potentially help develop diagnostic and prognostic liquid biopsy biomarkers that can improve pancreatic cancer patient outcomes.
In this study, we show that miR-483-3p is frequently overexpressed in noninvasive PanIN lesions as well as invasive PDAC, yet its expression is absent in almost all normal pancreatic ductal epithelia. Overexpression of miR-483-3p occurred as early as at the PanIN-1 lesion stage. Other dysregulated miRNAs previously reported in pancreatic cancer precursors37–39 have exhibited a similar pattern as miR-483-3p expression, which gradually increases as histological atypia of PanINs increases, while the putative miR-483-3p target gene SMAD4 protein expression is gradually lost.
PanINs are well-studied noninvasive precursors of PDAC. Although most PDAC lesions arise from PanINs, there are no effective approaches to detect these lesions because of their microscopic size. We previously reported that miR-483-3p expression was differentially upregulated in PDAC tissues as well as in a small subset of PanIN lesions when compared to normal pancreatic tissues.13 This study confirms our earlier findings in a large, independent set of cases where we again found increased miR-483-3p expression in PanIN lesions. Together these data suggest that miR-483-3p overexpression occurs as an early event in PDAC tumorigenesis, which may be an early, identifiable pancreatic cancer biomarker.
Over the last decade, large efforts have been made to study the expression pattern of circulating miRNAs in a variety of cancers.40–44 Circulating miRNAs have also become a central focus in the field of liquid biopsies as cell-free miRNAs are remarkably stable in various biofluids, including blood.45 In blood, circulating miRNAs are either encapsulated in extracellular vesicles (eg, exosomes) or combined with protein complexes (eg, argonaute-2 or high-density lipoprotein).46–49 Most studies have focused on total cell-free miRNAs in circulation, rather than the exosomal miRNAs yet recent studies have suggested that the majority of circulating miRNAs are concentrated in exosomes.50
We found that miR-483-3p was detectable in serum and serum exosomes from PDAC patients. miR-483-3p levels were significantly elevated in both serum and circulating exosome samples of PDAC patients compared to healthy controls. Comparatively, serum samples had a greater increase in miR-483-3p level compared to exosomes, with a better AUC value that could help discriminate PDAC patients from healthy subjects. We also found overexpression of miR-483-3p in PDAC tumor tissue which decreased in circulation after tumor resection, indicated that circulating miR-483-3p likely originates from PDAC cancer cells.
Serum miR-483-3p levels showed better diagnostic value for PDAC than circulating exosomal miR-483-3p, as supported by the relatively high AUC value of 0.81. The AUC value of serum miR-483-3p (0.81) in ROC curve was comparable to that of CA19-9 (0.87), a conventionally used tumor marker of PDAC. Combining serum and exosomal miR-483-3p levels increased the AUC value to 0.84. Serum miR-483-3p levels in early-stage PDAC (≤2 cm) could be used to distinguish these patients from healthy subjects, with AUC value of 0.83. Thus, circulating miR-483-3p has the potential to be utilized as a non-invasive liquid biomarker for early diagnosis of PDAC.
While serum miR-483-3p levels may serve as a PDAC diagnostic marker, high exosomal miR-483-3p expression was significantly correlated with increasing PDAC histological grade, high N-stage, and advanced overall clinical stage. These relationships may indicate that exosomal miR-483-3p expression is part of PDAC pathogenesis and the relationship with high N-stage may indicate that miR-483-3p contributes to PDAC metastatic potential. However, consistent with a previous study,51 we observed that serum miR-483-3p expression did not correlate with any clinicopathologic features.
Furthermore, our study suggested that serum exosomal miR-483-3p level could serve as a prognostic biomarker for PDAC as Kaplan–Meier analysis showed high levels of exosomal miR-483-3p in serum is correlated with significantly worse PDAC patient survival. Multivariable Cox regression analysis also showed that high exosomal miR-483-3p level in serum is an independent prognostic factor for PDAC patients.
miR-483-3p has been shown to promote cell proliferation and colony formation in pancreatic cancer cells by targeting SMAD4,15 a key tumor suppressor genes involved in pancreatic cancer pathogenesis.52 In this study, we found that miR-483-3p expression was inversely correlated with Smad4 protein expression in tissue samples of PDAC, consistent with a previous report.15 This inverse correlation was also identified in PanIN lesions. These observations suggest that miR-483-3p may exert oncogenic functions in the early pathogenesis of PDAC through down-regulation of SMAD4. Our observation that serum exosomal miR-483-3p levels were significantly associated with N stage led us to hypothesize that miR-483-3p may affect the migration and invasion potential of pancreatic tumor cells. We tested this in vitro and found that miR-483-3p promoted cell migration and invasion in transwell assays. These findings are consistent with known functions of the putative miR-483-3p target SMAD453,54 and further suggest that miR-483-3p may play a key role in PDAC progression.
There are two limitations in our studies. First, we have not included sera samples from chronic pancreatitis (CP) patients. CP is an important differential diagnosis of PDAC, which share similar clinical manifestation to PDAC. Second, the sample size is relatively small in our study.
In conclusion, we found that abnormal miR-483-3p overexpression occurs early in the premalignant PanIN lesions. We also provide evidence that serum miR-483-3p is a potential liquid biopsy marker for early detection of PDAC and serum exosomal miR-483-3p could potentially be used as a prognostic indicator of PDAC patient survival. Large-scale prospective multicenter studies are needed to further validate the potential of circulating miR-483-3p as PDAC biomarker.
Data Sharing Statement
The data used to support the findings of this study are available from the corresponding author upon request.
Acknowledgments
This work was supported by the National Science Foundations of China (Grant Nos.81400664, 81672648, 81472326, and 81341070), the CAMS Science and Technology Innovation Program Fund for Medical Sciences and Health (Grant No. 2016-I2M-1-001) and special funds from the CAMS Molecular Pathology Center and the Central Public Welfare Institutions of CAMS (Grant Nos. 2016ZX310176-4 and 2017PT31008).
Author Contributions
HS: experiments, data analysis and manuscript writing. YZ and JY: data extraction and analysis, figure and table construction. XB: sample collection and experiments. XF: technical support. XC and ZL: histopathological review. YW and LZ: critical reading and revision of the manuscript. SM: data analysis and interpretation. SY and JC: study conceiving and design, supervision, and result interpretation. All authors have given final approval for the submission of this manuscript and subsequently published versions. All authors contributed to data analysis, drafting or revising the article, have agreed on the journal to which the article will be submitted, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors declare no relevant conflicts of interest.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. doi:10.3322/caac.21590
3. Kamisawa T, Wood LD, Itoi T, Takaori K. Itoi T and Takaori K. Pancreatic cancer. Lancet. 2016;388(10039):73–85. doi:10.1016/S0140-6736(16)00141-0
4. Hruban RH, Goggins M, Parsons J, Kern SE. Parsons J and Kern SE. Progression model for pancreatic cancer. Clin Cancer Res. 2000;6(8):2969–2972.
5. Pittman ME, Rao R, Hruban RH. Classification, Morphology, Molecular Pathogenesis, and Outcome of Premalignant Lesions of the Pancreas. Arch Pathol Lab Med. 2017;141(12):1606–1614. doi:10.5858/arpa.2016-0426-RA
6. Hruban RH, Adsay NV, Albores–Saavedra J, et al. Klöppel G and Longnecker DS. Pancreatic intraepithelial neoplasia: a new nomenclature and classification system for pancreatic duct lesions. Am J Surg Pathol. 2001;25(5):579–586. doi:10.1097/00000478-200105000-00003
7. Hruban RH, Takaori K, Klimstra DS, et al. Fukushima N and Furukawa T. An illustrated consensus on the classification of pancreatic intraepithelial neoplasia and intraductal papillary mucinous neoplasms. Am J Surg Pathol. 2004;28(8):977–987. doi:10.1097/01.pas.0000126675.59108.80
8. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi:10.1016/S0092-8674(04)00045-5
9. Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857–866. doi:10.1038/nrc1997
10. Farazi TA, Spitzer JI, Morozov P, Tuschl T. Morozov P and Tuschl T. miRNAs in human cancer. J Pathol. 2011;223(2):102–115. doi:10.1002/path.2806
11. Bartel DP. MicroRNAs: target recognition and regulatory functions. cell. 2009;136(2):215–233. doi:10.1016/j.cell.2009.01.002
12. Zhang B, Pan X, Cobb GP, Anderson TA. Cobb GP and Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302(1):1–12. doi:10.1016/j.ydbio.2006.08.028
13. Wang CP, Sun Y, Wu HW, et al. Lu ZH and Chen J. Elevated miR-483-3p expression is an early event and indicates poor prognosis in pancreatic ductal adenocarcinoma. Tumor Biology. 2015;36(12):9447–9456. doi:10.1007/s13277-015-3690-x
14. Pepe F, Visone R, Veronese A. The Glucose-Regulated MiR-483-3p Influences Key Signaling Pathways in Cancer. Cancers. 2018;10(6):10. doi:10.3390/cancers10060181
15. Hao J, Zhang SY, Zhou YQ, Hu X, Shao C. Hu XG and Shao CH. MicroRNA 483-3p suppresses the expression of DPC4/Smad4 in pancreatic cancer. FEBS Lett. 2011;585(1):207–213. doi:10.1016/j.febslet.2010.11.039
16. Hessvik NP, Llorente A. Current knowledge on exosome biogenesis and release. Cell Mol Life Sci. 2018;75(2):193–208. doi:10.1007/s00018-017-2595-9
17. Vlassov AV, Magdaleno S, Setterquist R, Conrad R. Setterquist R and Conrad R. Exosomes: current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim Biophys Acta. 2012;1820(7):940–948. doi:10.1016/j.bbagen.2012.03.017
18. Becker A, Thakur BK, Weiss JM, Kim HS, Peinado H, Lyden D. Peinado H and Lyden D. Extracellular Vesicles in Cancer: cell-to-Cell Mediators of Metastasis. Cancer Cell. 2016;30(6):836–848. doi:10.1016/j.ccell.2016.10.009
19. Maas SLN, Breakefield XO, Weaver AM. Breakefield XO and Weaver AM. Extracellular Vesicles: unique Intercellular Delivery Vehicles. Trends Cell Biol. 2017;27(3):172–188. doi:10.1016/j.tcb.2016.11.003
20. Zhang J, Li S, Li L, Li M, Guo C. Yao J and Mi S. Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics Proteomics Bioinformatics. 2015;13:17–24. doi:10.1016/j.gpb.2015.02.001
21. Yang H, Zhang H, Ge S, et al. Ying G and Ba Y. Exosome-Derived miR-130a Activates Angiogenesis in Gastric Cancer by Targeting C-MYB in Vascular Endothelial Cells. Mol Ther. 2018;26(10):2466–2475. doi:10.1016/j.ymthe.2018.07.023
22. Dong L, Pu Y, Zhang L, et al. Chen X and Su C. Human umbilical cord mesenchymal stem cell-derived extracellular vesicles promote lung adenocarcinoma growth by transferring miR-410. Cell Death Dis. 2018;9(2):218. doi:10.1038/s41419-018-0323-5
23. Fong MY, Zhou W, Liu L, et al. Chen Y and Wang SE. Breast-cancer-secreted miR-122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nat Cell Biol. 2015;17:183–194. doi:10.1038/ncb3094
24. Fang T, Lv H, Lv G, et al. Yang W and Wang H. Tumor-derived exosomal miR-1247-3p induces cancer-associated fibroblast activation to foster lung metastasis of liver cancer. Nat Commun. 2018;9(1):191. doi:10.1038/s41467-017-02583-0
25. Shao Y, Chen T, Zheng X, et al. Shao J and Zheng S. Colorectal cancer-derived small extracellular vesicles establish an inflammatory premetastatic niche in liver metastasis. Carcinogenesis. 2018;39(11):1368–1379. doi:10.1093/carcin/bgy115
26. Tominaga N, Kosaka N, Ono M, et al. Nakagama H and Ochiya T. Brain metastatic cancer cells release microRNA-181c-containing extracellular vesicles capable of destructing blood-brain barrier. Nat Commun. 2015;6:6716. doi:10.1038/ncomms7716
27. Matsumura T, Sugimachi K, Iinuma H, et al. Ochiya T and Mimori K. Exosomal microRNA in serum is a novel biomarker of recurrence in human colorectal cancer. Br J Cancer. 2015;113(2):275–281. doi:10.1038/bjc.2015.201
28. Reese M, Flammang I. Yang Z and Dhayat SA. Potential of Exosomal microRNA-200b as Liquid Biopsy Marker in Pancreatic Ductal Adenocarcinoma. Cancers. 2020;12.
29. Bhagirath D, Yang TL, Bucay N, et al. Tanaka Y and Saini S. microRNA-1246 Is an Exosomal Biomarker for Aggressive Prostate Cancer. Cancer Res. 2018;78(7):1833–1844. doi:10.1158/0008-5472.CAN-17-2069
30. Amin M, Edge S, Greene F, et al. AJCC Cancer Staging Manual. New York: Springer; 2017.
31. Wilentz RE, Su GH, Le Dai J, et al. Kern SE and Hruban RH. Immunohistochemical labeling for dpc4 mirrors genetic status in pancreatic adenocarcinomas: a new marker of DPC4 inactivation. Am J Pathol. 2000;156(1):37–43. doi:10.1016/S0002-9440(10)64703-7
32. Li Y, Kowdley KV. Method for microRNA isolation from clinical serum samples. Anal Biochem. 2012;431(1):69–75. doi:10.1016/j.ab.2012.09.007
33. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi:10.1006/meth.2001.1262
34. Shen W, Han Y, Huang B, et al. MicroRNA-483-3p Inhibits Extracellular Matrix Production by Targeting Smad4 in Human Trabecular Meshwork Cells. Invest Ophthalmol Vis Sci. 2015;56(13):8419–8427. doi:10.1167/iovs.15-18036
35. Anderson BA, McAlinden A. miR-483 targets SMAD4 to suppress chondrogenic differentiation of human mesenchymal stem cells. J Orthop Res. 2017;35:2369–2377. doi:10.1002/jor.23552
36. Nevala-Plagemann C, Hidalgo M, Garrido-Laguna I. From state-of-the-art treatments to novel therapies for advanced-stage pancreatic cancer. Nat Rev Clin Oncol. 2020;17(2):108–123. doi:10.1038/s41571-019-0281-6
37. Habbe N, Koorstra JBM, Mendell JT, et al. Hong SM and Maitra A. MicroRNA miR-155 is a biomarker of early pancreatic neoplasia. Cancer Biol Ther. 2009;8:340–346. doi:10.4161/cbt.8.4.7338
38. Ryu JK, Hong SM, Karikari CA, Hruban RH, Goggins MG, Maitra A. Aberrant MicroRNA-155 Expression Is an Early Event in the Multistep Progression of Pancreatic Adenocarcinoma. Pancreatology. 2010;10:66–73. doi:10.1159/000231984
39. Du Rieu MC, Torrisani J, Selves J. MicroRNA-21 Is Induced Early in Pancreatic Ductal Adenocarcinoma Precursor Lesions. Clin Chem. 2010;56(4):603–612. doi:10.1373/clinchem.2009.137364
40. Lin XJ, Chong Y, Guo ZW, et al. Zheng L and Zhuang SM. A serum microRNA classifier for early detection of hepatocellular carcinoma: a multicentre, retrospective, longitudinal biomarker identification study with a nested case-control study. Lancet Oncol. 2015;16:804–815. doi:10.1016/S1470-2045(15)00048-0
41. Hur K, Toiyama Y, Okugawa Y, Ide S, Imaoka H. Boland CR and Goel A. Circulating microRNA-203 predicts prognosis and metastasis in human colorectal cancer. Gut. 2017;66(4):654–665. doi:10.1136/gutjnl-2014-308737
42. Schultz NA, Dehlendorff C, Jensen BV, et al. Andersen KK and Johansen JS. MicroRNA biomarkers in whole blood for detection of pancreatic cancer. JAMA. 2014;311(4):392–404. doi:10.1001/jama.2013.284664
43. Liu H, Zhu L, Liu B, Yang L, Meng X, Zhang W. Ma Y and Xiao H. Genome-wide microRNA profiles identify miR-378 as a serum biomarker for early detection of gastric cancer. Cancer Lett. 2012;316(2):196–203. doi:10.1016/j.canlet.2011.10.034
44. Foss KM, Sima C, Ugolini D, Neri M, Allen KE, Weiss GJ. Allen KE and Weiss GJ. miR-1254 and miR-574-5p: serum-based microRNA biomarkers for early-stage non-small cell lung cancer. J Thorac Oncol. 2011;6(3):482–488. doi:10.1097/JTO.0b013e318208c785
45. Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci. 2008;105(30):10513–10518. doi:10.1073/pnas.0804549105
46. Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Lee JJ and Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654. doi:10.1038/ncb1596
47. Hunter MP, Ismail N, Zhang X, et al. Jarjoura D and Marsh CB. Detection of microRNA expression in human peripheral blood microvesicles. PLoS One. 2008;3:e3694. doi:10.1371/journal.pone.0003694
48. Arroyo JD, Chevillet JR, Kroh EM, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011;108(12):5003–5008. doi:10.1073/pnas.1019055108
49. Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. Shamburek RD and Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13(4):423–433. doi:10.1038/ncb2210
50. Gallo A, Tandon M, Alevizos I, Illei GG. Alevizos I and Illei GG. The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PLoS One. 2012;7(3):e30679. doi:10.1371/journal.pone.0030679
51. Abue M, Yokoyama M, Shibuya R, et al. Sugamura K and Satoh K. Circulating miR-483-3p and miR-21 is highly expressed in plasma of pancreatic cancer. Int J Oncol. 2015;46(2):539–547. doi:10.3892/ijo.2014.2743
52. Makohon-Moore A, Iacobuzio-Donahue CA. Pancreatic cancer biology and genetics from an evolutionary perspective. Nat Rev Cancer. 2016;16(9):553–565. doi:10.1038/nrc.2016.66
53. Wang Z, Li Y, Zhan S, et al. Liu S and Xing X. SMAD4 Y353C promotes the progression of PDAC. BMC Cancer. 2019;19(1):1037. doi:10.1186/s12885-019-6251-7
54. Zhao M. Mishra L and Deng CX. The role of TGF-beta/SMAD4 signaling in cancer. Int J Biol Sci. 2018;14:111–123. doi:10.7150/ijbs.23230
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.