Back to Journals » OncoTargets and Therapy » Volume 13
Up-Regulation of FSTL3, Regulated by lncRNA DSCAM-AS1/miR-122-5p Axis, Promotes Proliferation and Migration of Non-Small Cell Lung Cancer Cells
Authors Gao L, Chen X , Wang Y, Zhang J
Received 28 October 2019
Accepted for publication 21 February 2020
Published 1 April 2020 Volume 2020:13 Pages 2725—2738
DOI https://doi.org/10.2147/OTT.S236359
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr XuYu Yang
Liang Gao,1,2 Xiaochen Chen,1,2 Yongxiang Wang,3 Jianbin Zhang1,2
1Department of Oncology, Zhejiang Provincial People’s Hospital, Hangzhou 310022, Zhejiang Province, People’s Republic of China; 2Department of Oncology, People’s Hospital of Hangzhou Medical College, Hangzhou 310014, Zhejiang Province, People’s Republic of China; 3Department of Abdominal Surgery, Zhejiang Cancer Hospital, Hangzhou 310022, Zhejiang Province, People’s Republic of China
Correspondence: Xiaochen Chen
Department of Oncology, Zhejiang Provincial People’s Hospital, Hangzhou 310022, People’s Republic of China
Tel +86 57185893183
Email [email protected]
Background: Follistatin-like 3 (FSTL3) binds and inactivates activin, a growth factor with cell growth and differentiation. Previous studies reported that it is overexpressed in invasive breast cancers, and its expression and function in non-small cell lung cancer (NSCLC) remain unclear.
Materials and Methods: Immunohistochemistry was employed to probe the expression of FSTL3 in NSCLC tissues. Real-time PCR (RT-PCR) was applied to detect the expression of lncRNA DSCAM-AS1 and miR-122-5p. A549 cells and H1299 cells were used as cell models. The biological influence of FSTL3 on cells was studied using CCK-8 assay, wound healing assay and transwell assay in vitro, respectively. In vivo subcutaneous xenotransplanted tumor model and tail vein injection model in mice were also constructed to validate the roles of FSTL3. Interactions between miR-122-5p and FSTL3, DSCAM-AS1 and miR-122-5p were determined by bioinformatics analysis, RT-PCR, and dual-luciferase reporter assay.
Results: FSTL3 and DSCAM-AS1 were remarkably up-regulated in NSCLC samples, and miR-122-5p was down-regulated. FSTL3 was associated with worse prognosis of NSCLC patients. FSTL3 knockdown markedly inhibited the viability, migration and invasion of NSCLCs in vitro and in vivo. DSCAM-AS1 could down-regulate miR-122-5p via sponging it, and FSTL3 was a target gene of miR-122-5p.
Conclusion: Taken together, our study identified that FSTL3 was a new oncogene of NSCLC, which was regulated by DSCAM-AS1 and miR-122-5p. These findings suggested that FSTL3, DSCAM-AS1 and miR-122-5p might serve as a new valuable therapeutic target for NSCLC.
Keywords: FSTL3, NSCLC, lncRNA DSCAM-AS1, miR-122-5p, proliferation and migration
Introduction
Lung cancer is the most common tumors with the highest morbidity and mortality in the world. Non-small cell lung cancer (NSCLC) accounts for about 85% of all lung cancer cases.1 NSCLC often has no obvious clinical manifestations at the early stage, leading to the difficulty in early diagnosis. In addition, lymph node and distant organ metastasis are very likely to occur in NSCLC.2 In recent years, although great progress has been made in diagnosis and treatment, the 5-year survival rate of NSCLC patients is still less than 15%.3 Therefore, it is of great significance to clarify the mechanism of the occurrence and progression of NSCLC, and to find novel therapy targets.
Follistatin-like 3 (FSTL3) (also FSRP or FLRG), is a member of the FSTL family.4 FSTL3 is a novel cytokine that regulates insulin sensitivity and counteracts activin or myostatin signaling. Recent study shows that FSTL3 is involved in regulating glucose and fat homeostasis,5 testicular aging and testicular size,6 bone remodeling,7 osteoarthritis8 and other physiological process or diseases. Additionally, FSTL3 has also been found to regulate the progression of tumors. For example, FSTL3 is overexpressed in invasive breast cancer and can promote the proliferation of tumor cells by antagonizing endogenous activators.9 However, its expression and biological function in NSCLC are unknown to date.
Long non-coding RNA (lncRNA) is a kind of non-coding RNA with a length of over 200 nucleotides and has no protein-coding function. However, it can regulate protein expression through epigenetics, transcription and post-transcriptional level.10 Previous studies have shown that abnormal lncRNA expression has impacts on the occurrence and progression of NSCLC.11,12 A recent study reported that DSCAM-AS1 can act as an oncogene in NSCLC by targeting BCL11A, and DSCAM-AS1 overexpression can promote proliferation and migration of NSCLC cells.13 Among non-coding RNA molecules, in addition to lncRNA, there is also a kind of non-coding RNA molecules – microRNA (miRNA), a class of highly conserved short non-coding RNA molecules with a length of about 22 nucleotides, which have important biological regulation capabilities.14 As a member of miRNAs, miR-122-5p has abnormal expression in tumors and is of great importance in the occurrence and progression of tumors. A previous study has proven that miR-122-5p is down-regulated in gastric cancer tissues and cells and inhibits the migration and invasion of gastric cancer cells by regulating DUSP4.15 Additionally, miR-122-5p is under-expressed in NSCLC, which can regulate the epithelial–mesenchymal transition (EMT) of NSCLC cells.16,17
This study explored the biological function of FSTL3 in NSCLC. We proved that FSTL3 expression was up-regulated in NSCLC, and overexpression of FSTL3 could promote proliferation and metastasis of NSCLC, while knockdown of FSTL3 played an opposite role. In addition, we also investigated the mechanism of dysregulation of FSTL3 in NSCLC. We demonstrated a competing endogenous mechanism that FSTL3 was a target gene of miR-122-5p, and could be indirectly regulated by DSCAM-AS1.
Materials and Methods
Tissue Samples
Sixty patients with NSCLC who underwent surgical treatment in our hospital from 2015 to 2018 were enrolled. Cancer tissue samples and adjacent normal tissues were taken for pathological examination after the operation. The age of patients ranged from 26 to 70 years, including 28 males and 32 females. All patients involved gave informed consent to the study and signed a written consent form, and that this was conducted in accordance with the Declaration of Helsinki. All specimens were immediately removed and stored in liquid nitrogen at −196°C for RNA extraction and other experiments. The collection and use of patient tissue samples were approved by the Ethics Committee of Zhejiang Provincial People’s Hospital. All procedures in the protocol were in compliance with the guidelines of the Animal Welfare Act, the Guide for the Care and Use of Laboratory Animals, and the Office of Laboratory Welfare.
Cell Culture
The normal bronchial cell line 16HBE and NSCLC cell lines (A549, NCI-H460, H1299, L9981 and NCI-H292) were purchased from Cell center of Chinese Academy of Sciences (Shanghai, China). The cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM, Gibco, Grand Island, New York) supplemented with 10% fetal bovine serum (FBS; Gibco, Life Technologies, Carlsbad, CA) in a constant temperature incubator at 37°C and in 5% CO2 and saturated humidity.
Cell Transfection
FSTL3 overexpression plasmids, sh-FSTL3 lentivirus infection, DSCAM-AS1 overexpression plasmids, miR-122-5p mimics or inhibitors, and corresponding negative control groups were all constructed by Shanghai Jikai Genochemical Technology Co., Ltd. (Genechem, Shanghai, China). Human lung cancer cells A549 and H1299 in logarithmic growth phase were selected and trypsinized to adjust the cell concentration to 1 × 108/L. 0.5 mL was inoculated in a 24-well culture plate at 37°C and cultured in a constant temperature incubator until the cell adhesion coverage rate reached 70~90%. The cells were washed in serum-free medium for 2 times and then washed in DMEM medium for 2 times. 400 μL serum-free DMEM medium was added to continue culture. Subsequently, Lipofectamine 3000 (Invitrogen; ThermoFisher Science, Inc.) was applied according to the supplier’s instructions. The recombinant plasmid was transfected into A549 cells, H1299 cells and HEK293T cells. The transfection efficiency was detected by qRT-PCR. The cells were incubated at 37°C and 5% CO2 for 24 hrs, pending further analysis.
Immunohistochemistry (IHC)
Paraffin sections were dewaxed routinely and hydrated with gradient ethanol. Sections were xylene dewaxed, dehydrated and rehydrated. Then, the sections were incubated with primary antibody Anti-FSTL3 (Abcam, ab86055, 1:2000) overnight and secondary antibody for 30 mins at room temperature, respectively. Subsequently, the sections were rinsed thoroughly with PBS solution. Following that, DAB (Beijing Airan Biotechnology Co., Ltd.) was employed to terminate the reaction after the color was developed. The scoring standard of immunohistochemistry was completed by the pathologists from the Department of Pathology in our hospital.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA was extracted from A549 and H1299 lung cancer cells by TRIzol method, and genomic DNA was removed by deoxyribonuclease I. According to the operation procedure of Taqman® reverse transcription kit (Thermo Fisher, Shanghai, China), RNA was reverse-transcribed to obtain cDNA. After the reaction, PCR was used to further amplify the cDNA with SYBR Green Master Mix (Roche, Shanghai, China). Primer sequences of each molecule were as follows: FSTL3 positive primer: 5ʹ-CTGGGATCCTGAGCACGTAT-3ʹ. Reverse primer: 3ʹ-GCCAGGGTCCAATGTTTCTA-5ʹ. miR-122-5p forward primer: 5 ‘-GGGGTGGAGTGTGACAATG-3ʹ. Reverse primer: 5ʹ-CAGTGCGTGTCGTGGAGT-3ʹ. DSCAM-AS1 forward primer: 5ʹ‐CCTATCCCTTTCTCTAAGAA‐3ʹ; Reverse primer: 5ʹ‐ACTTCTGCAAAAACGTGCTG‐3ʹ. GAPDH forward primer: 5ʹ-CCAGGGCTGCTTTTAACTCT-3ʹ; Reverse primer: 5ʹ-GGACTCCACGACGTACTCA-3ʹ. U6 forward primer: 5ʹ-GCTCGCTTCGGCAGCCACA-3ʹ; Reverse primer: 5ʹ-AACGCTTCACGAATTTGCGT-3ʹ. The relative expressions of the genes were measured with 2−ΔΔCt method.
CCK-8 Method
A549 and H1299 cells in logarithmic growth phase were taken and trypsinized with trypsin. After the cell density was adjusted to 2 × 104/mL, cells were inoculated into 96-well plate with 100 μL cell suspension per well. After that, 96-well plate was placed in incubator for further culture. After 24 hrs, 10 μL CCK8 kit (Beyotime Biotechnology, Hangzhou, China) was added and incubated in an incubator for 1 hr. After termination of culture, 96-well plates were placed in an microplate reader to determine the optical density (OD) value of each well at 450 nm wavelength. Similarly, the OD value of cells was measured at 48, 72 and 96 hrs, respectively.
Immunofluorescence
Cells were inoculated into the 24-well plates. 200 μL 5 μmol/L Edu solution was added to each well, the cells were incubated for 2 hrs and then washed with PBS. After that, the cells were fixed with paraformaldehyde and incubated at room temperature for 10 mins. Afterwards, 200 μL 2mg/mL Glycine was added and incubated for 5 mins and washed with PBS on a shaking bed for 5 mins. Then, 100 μL PBS containing 0.5% TritonX-100 was added into each well for 10 mins. After that, Cell-Light™ EdU Apollo®488 In Vitro Imaging Kit (RiboBio, Guangzhou, China) was used to stain the cells in dark for 30 mins, and 1 × Hoechst 33342 DNA-staining solution (Beyotime Biotechnology, Hangzhou, China) was added and incubated at room temperature in dark for 20 mins. After that, cells were washed with PBS and the fluorescent signal was observed under a fluorescence microscope.
Transwell Assay
Transwell assay was used to detect the migration A549 and H1299 cells. 2 × 104 cells suspended in serum-free medium were seeded into Transwell chamber’ upper chamber, and 600 µL medium containing 20% FBS were added into the lower chamber, and cultured at 37°C. After 12 hrs, the cells failing to migrate were removed, and the migrated cells were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet. After washing and drying, the migrated cells were photographed and counted.
Luciferase Reporter Assay
Luciferase reporter gene assay was conducted to verify the targeting between miR-122-5p and DSCAM-AS1 or 3′-UTR of FSTL3. The wild type (WT) DSCAM-AS1 sequence or the WT 3ʹ-UTR fragment from FSTL mRNA including the predicted binding site of miR-122-5p were amplified and inserted into the pmirGLO dual-luciferase vector (Promega, Madison, WI, USA) to construct the report vector pmirGLO-DSCAM-AS1-WT or pmirGLO-FSTL3-WT. GeneArt™ Site-Directed Mutagenesis PLUS System (cat. no. A14604, Thermo Fisher Scientific, Inc.) was used to mutate the presumed binding site of miR-122-5p in DSCAM-AS1 or FSTL3 3ʹ-UTR. Mutant (MUT) DSCAM-AS1 or FSTL3 3′-UTR were inserted into pmirGLO vector to form report vector pmirGLO-DSCAM-AS1-Mut or pmirGLO-FSTL3-Mut. The corresponding reporter vectors and miR-122-5p or NC mimics were co-transfected into HEK293T cells and incubated for 48 hrs. Luciferase activity was then measured using Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA).
RNA-Binding Protein Immunoprecipitation
RIP experiments were conducted according to the instructions of the manufacturer of Magna RNA-binding protein immunoprecipitation kit (Millipore, Life Science, Shanghai, China). The cultured cells were pre-lysed to obtain whole-cell extracts, and the supernatants were collected for immunoprecipitation by adding the cleavage buffer containing protease inhibitors and RNase enzyme inhibitors. The lysate was incubated overnight with protein A Sepharose magnetic beads (8 μ g, MA5-23515, Invitrogen) combined with anti-Ago2 antibody at 4°C. Next, the magnetic beads were washed and protease K was used to separate the immunoprecipitated RNA. Anti-IgG (ab6715, Abcam, Cambridge, UK) was also used as control. Then, qRT-PCR was conducted to detect the enrichment of the molecules.
Western Blot Method
Cells were added with RIPA lysate containing 1% PMSF to extract the total protein. After sodium dodecyl sulfate-polyacrylamide gel electrophoresis, the protein samples were separated. After that, protein was electrotransferred to polyvinylidene fluoride membrane. Then, the membrane was blocked with 5% skim milk at room temperature for 1 hrs, washed with TBST for 3 times, and then incubated overnight with primary antibodies including anti-FSTL3 (Abcam, ab86055, 1:1000) and anti-GAPDH (Abcam, ab8245, 1:3000) at 4°C. After washing with TBST again, the membrane was incubated with secondary antibody labeled with HRP for 1 hr at room temperature. Then, TBST was used to wash the membranes for 3 times. Ultimately, ECL reagent (Millipore, Shanghai, China) was used to show the bands.
Nude Mice Study
All animal experiments were approved by the Animal Care and Use Committee of Zhejiang Provincial People’s Hospital. Female BALB/c nude mice (4 weeks old) were purchased from Topbiotech Biotechnology Co., Ltd. (Shenzhen, China). Mice were randomly divided into sh-NC group and sh-FSTL3 group with 10 mice in each group. NSCLCs (1 × 107 cells) of the two groups were subcutaneously injected into the back of mice, respectively, and the volume of tumors was monitored every 7 days. On the 35th day after injection, mice were sacrificed, xenografted tumors were removed and weighed. In the lung metastasis study, 1×107 cells in the two groups were injected into caudal vein of 10 mice, respectively. Two weeks later, mice were sacrificed and lung metastasis was evaluated by hematoxylin-eosin (HE) staining.
Statistical Analysis
SPSS 17.0 statistical software (SPSS Inc., Chicago, IL, USA) was used to analyze the data. The measurement data were expressed as mean ± standard deviation (x±s). Student’s t-test was used for comparison between two groups. The data of counting data were expressed by four-grid table (or percentage), and the difference between the two groups was analyzed by χ2. The difference was statistically significant with P < 0.05.
Results
The Expression of FSTL 3 Were Up-Regulated in NSCLC
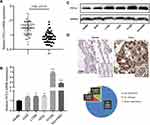
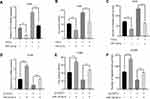
To preliminarily explore the expression characteristics of FSTL3 in NSCLC tissues, we used qRT-PCR to detect the expression of FSTL3 mRNA in NSCLC tissues and adjacent non-cancerous lung tissues. As shown, FSTL3 was significantly up-regulated in NSCLC tissues (Figure 1A). In addition, the expression of FSTL3 in NSCLC cell lines was detected by qRT-PCR and Western blot. It showed the levels of FSTL3 mRNA and protein in NSCLC cell lines were significantly higher than those in 16HBE cells (Figure 1B and C). Subsequently, we used IHC to examine FSTL3 expression in 60 pairs of NSCLC tissues and corresponding non-cancerous lung tissues. As shown, FSTL3 expression was up-regulated in most NSCLC patients (75%, 45/60) (Figure 1D). These results implied the cancer-promoting effect of FSTL3 in NSCLC.
FSTL3 Expression Was Correlated with Multiple Clinicopathological Features and Survival Rate of NSCLC Patients
To clarify the role of FSTL3 in the occurrence and progression of NSCLC, we then used the above-mentioned 60 NSCLC samples to analyze the correlation between FSTL3 expression and various pathological indicators of NSCLC patients (Table 1). Chi-square test indicated that high expression of FSTL3 in tumor tissues was significantly correlated with local lymph node invasion (P=0.0395) and increased T staging (P=0.0020) in NSCLC patients, but not significantly correlated with age, gender, smoking history, tumor type and tumor differentiation (P>0.05). In addition, Kaplan-Meier analysis was performed using TCGA data with online database Gepia (http://gepia.cancer-pku.cn/), and we demonstrated that the overall survival time and disease-free survival time of patients (both adenocarcinoma and squamous carcinoma) with higher FSTL3 expression were shorter than those with lower FSTL3 expression (Figure 2A–D). These outcomes implied that FSTL3 may promote the occurrence and metastasis of NSCLC.
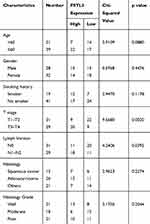 |
Table 1 Relationship Between FSTL3 Levels and Clinical Characteristics of NSCLC (N=60) |
FSTL3 Regulated NSCLC Cell Proliferation and Metastasis in vitro
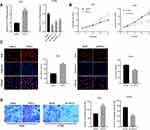
After FSTL3 was detected to be significantly up-regulated in NSCLC tissues and cell lines, we will explore its function in NSCLC cells. H1299 and A549 cell lines were selected and we successfully construct FSTL3 knockdown model and overexpression model, respectively (Figure 3A). On this basis, the proliferation ability of the above cells was tested by CCK-8 assay and Edu assay. The proliferation ability of the FSTL3 knockdown group was significantly impeded compared with the sh-NC group in H1299 cells. On the contrary, FSTL3 overexpression facilitated the proliferation of A549 cells (Figure 3B and C). Additionally, we tested the effect of FSTL3 on cell metastasis by Transwell experiment. The results showed that compared with control group, FSTL3 over expression significantly promoted the migration of A549 cells. Meanwhile, compared with control group, the migration of H1299 cells with FSTL3 knocked down was significantly reduced (Figure 3D). Collectively, these data indicated that FSTL3 could promote the malignant phenotypes of NSCLC cells.
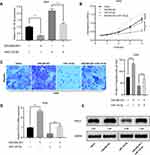
Inhibition of FSTL3 Could Inhibit Tumor Growth and Metastasis in vivo
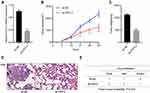
To work out whether FSTL3 affects tumor growth and metastasis in vivo, we constructed stable cell lines with low expression of FSTL3 using lentiviral vectors system (Figure 4A). On this basis, a subcutaneous tumor model was established with nude mice. As shown, the volume of tumors formed by H1299/sh-FSTL3 cells was smaller than that formed by H1299/sh-NC cells (Figure 4B). Consistently, after the mice were sacrificed, the weight of tumors in H1299/sh-FSTL3 group was remarkably lower than H1299/sh-NC cells (Figure 4C). To further determine the effect of FSTL3 knockdown on tumor metastasis in vivo, H1299/sh-FSTL3 and H1299/sh-NC cells were transplanted into the lateral caudal vein of nude mice, respectively. Two weeks later, mice were killed and lung metastases were examined by HE stain. As shown, the number of lung metastases in mice injected with H1299/sh-FSTL3 cells decreased significantly (Figure 4D). Eight out of 10 mice in the control group showed severe lung metastasis, the incidence of which was significantly higher than that of H1299/sh-NC group (1/10) (Figure 4E). These results further validated FSTL3 exerted an important role in the growth and metastasis of NSCLC.
FSTL3 Was a Target Gene of miR-122-5p
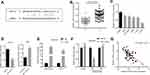
In order to elucidate the upstream mechanism of FSTL3 regulating NSCLC phenotypes, we conducted bioinformatics analysis through the StarBase database (http://www.starbase.sysu.edu.cn/). The data showed that FSTL3 contained target sites of miR-122-5p (Figure 5A). We measured the expression of miR-122-5p in NSCLC tissues and cell lines. We found that the expression of miR-122-5p in NSCLC tissues and cell lines was significantly lower than that in normal tissues and cells (Figure 5B and C). To further verify whether miR-122-5p was indeed a target of FSTL3, we transfected miR-122-5p mimics and inhibitors into H1299 and A549 cell lines, respectively, and qRT-PCR showed that the expression level of FSTL3 mRNA in H1299 cells and A549 cells were down-regulated and up-regulated, respectively (Figure 5D). RIP assay showed that the miR-122-5p coexisted with FSTL3 in NSCLC (Figure 5E). In addition, the activity of luciferase of wild type pGL3-FSTL3 vector was decreased by miR-122-5p mimics, but the effect of miR-122-5p mimics on mutant pGL3-FSTL3 vector was not significant (Figure 5F). We concluded that miR-122-5p could bind directly to FSTL3 mRNA at the recognition site. Importantly, we also demonstrated that the expression of FSTL3 in NSCLC samples was negatively correlated with the expression level of miR-122-5p (Figure 5G). These results supported that FSTL3 could be negatively regulated by miR-122-5p.
miR-122-5p Reversed the Effects on NSCLC Cells Induced by FSTL3 Overexpression
In order to further verify the interaction between FSTL3 and miR-122-5p, and to explore the importance of miR-122-5p in regulating NSCLC progression, we overexpressed miR-122-5p in NSCLCs stably overexpressing FSTL3 (Figure 6A). We found that miR-122-5p overexpression significantly impeded cell proliferation and migration compared to the control group. Compared with the FSTL3 group, the overexpression of miR-122-5p weakened the effect of overexpression of FSTL3 on the proliferation and migration of NSCLC cells (Figure 6B and C). In addition, miR-122-5p inhibitors promoted the expression of FSTL3 (Figure 6D). miR-122-5p inhibitors also promoted cell proliferation and migration compared to the control group; compared with the FSTL3 knockdown group, inhibition of miR-122-5p weakened the effect of knockdown of FSTL3 on proliferation and migration of NSCLC CELLS (Figure 6E and F).
DSCAM-AS1 Functioned as ceRNA to Regulate FSTL3 by Targeting miR-122-5p
As mentioned above, it was confirmed that the expression of FSTL3 could be regulated by miR-122-5p. Then, we tried to further explore the upstream regulatory mechanism of miR-122-5p. Bioinformatics prediction indicated that DSCAM-AS1 could probably target miR-122-5p (Figure 7A). By qRT-PCR, we found that DSCAM-AS1 was negatively correlated with the expression of miR-122-5p, and DSCAM-AS1 was positively correlated with the expression of FSTL3 (Figure 7B and C). RIP assay also showed that DSCAM-AS1 coexisted with miR-122-5p in NSCLC (Figure 7D). In addition, the luciferase activity of wild type pGL3-DSCAM-AS1 vector was decreased by miR-122-5p, but the effect of miR-122-5p on mutant pGL3-DSCAM-AS1 vector was not significant (Figure 7E). Besides, DSCAM-AS1 was also up-regulated in NSCLC cell lines, especially in H1299 cell line (Figure 7F).
miR-122-5p Reversed the Proliferation and Metastasis of NSCLCs Induced by DSCAM-AS1
We also observed that overexpression of DSCAM-AS1 significantly reduced the expression of miR-122-5p (Figure 8A). Moreover, the results of CCK-8 assay showed that the proliferation of cells inhibited by miR-122-5p mimics was reversed by DSCAM-AS1 overexpression (Figure 8B). Similarly, Transwell assay showed that DSCAM-AS1 attenuated the inhibitory effect of miR-122-5p on cell migration (Figure 8C). Subsequently, we detected the expression of FSTL3 in cells overexpressing DSCAM-AS1 or/and miR-122-5p. Compared with the control group, the overexpression of DSCAM-AS1 significantly increased the expression of FSTL3 mRNA and protein, and the overexpression of DSCAM-AS1 and miR-122-5p significantly increased the expression of FSTL3 mRNA and protein compared with the miR-122-5p group (Figure 8D and E). These results further validated the regulatory function of DSCAM-AS1 on miR-122-5p and FSTL3.
Discussion
With the continuous exploration of the pathogenesis of NSCLC, more and more molecules have been found to be enrolled in the occurrence and progression of this disease.18,19 Previous studies have found that FSTL family members have abnormal expression in tumors and are involved in the tumorigenesis and cancer progression. For example, FSTL1 expression is down-regulated in NSCLC, and its overexpression can significantly inhibit cell proliferation, migration and invasion, and induce apoptosis,20 while FSTL1 expression is up-regulated in hepatocellular carcinoma, and knocking down its expression can promote apoptosis and inhibit cell proliferation.21 A study reports that FSTL5 expression is down-regulated in hepatocellular carcinoma, and overexpression can inhibit cancer cell viability.22 As a member of FSTL family, FSTL3 also has abnormal expression and matters in tumor. For example, FSTL3 may serve as a cancer biomarker in breast and hepatocellular carcinoma.23,24 FSTL3 is up-regulated in breast cancer, and overexpression can promote the occurrence and progression of tumor.9,25 Herein, we found for the first time that FSTL3 was up-regulated in NSCLC, and its high expression level was significantly correlated with poor prognosis in NSCLC patients. In addition, we also found that knockdown of its expression could markedly impede the proliferation, migration and invasion of NSCLC cells, while overexpression of FSTL3 played a cancer-promoting effect. These results indicated that FSTL3, as an oncogene, was involved in the occurrence and progression of NSCLC.
miRNAs can bound to a specific mRNA to regulate gene expression. The dysregulation of miRNAs was involved in cancer biology. For example, miR-21 can promote the proliferation and migration of breast cancer via targeting LZTFL1,26 and miR-221 can promote the proliferation of cutaneous squamous cell carcinoma by targeting PTEN.27 In NSCLC, the expression of miR-221 and miR-10a were up-regulated, and their overexpression could accelerate the proliferation, migration and invasion of cancer cells.28,29 Consistent with previous reports,16,17 we confirmed that miR-122-5p was down-regulated in NSCLC tissues and involved in the progression of NSCLC as a tumor suppressor. In addition, we also found that there was a binding site between miR-122-5p and the 3ʹUTR of FSTL3, and the expression of FSTL3 and miR-122-5p was negatively correlated in NSCLC tissues. The expression of miR-122-5p in FSTL3 overexpressed cells was decreased, while the expression level of miR-122-5p in FSTL3 knockdown cells was increased, and the overexpression of miR-122-5p weakened the effect of FSTL3 overexpression on malignant phenotypes of NSCLCs.
LncRNA can regulate the expression of miRNAs as ceRNA and affect the level of downstream proteins. In NSCLC, lncRNA NEAT1 can promote the progression of NSCLC by competitively binding to miR-377-3p to affect the expression of E2F3;30 lncRNA SNHG1 promotes the progression of NSCLC by inhibition miR-101-3p and regulating Wnt/β-catenin signaling pathway.31 Recent studies have shown that DSCAM-AS1 is abnormally expressed in a variety of tumors and can play an important role by regulating downstream target genes. DSCAM-AS1 is overexpressed in breast cancer, which can promote tumor growth by inhibiting miR-204-5p and up-regulating RRM2.32 It is also up-regulated in hepatocellular carcinoma, which can promote the progression of hepatocellular carcinoma through sponging miR-338-3p.33 In addition, a study has shown that DSCAM-AS1 is overexpressed in NSCLC and can act as an oncogenic lncRNA by regulating BCL11A.13 In this study, we found that DSCAM-AS1 could target miR-122-5p. We also found that DSCAM-AS1 knockdown significantly reduced FSTL3 mRNA and protein levels, while DSCAM-AS1 overexpression increased FSTL3 mRNA and protein levels, and DSCAM-AS1 expression was positively correlated with FSTL3 expression in NSCLC tissues. These results, at least partially, explained the mechanism of FSTL3 dysregulation in NSCLC.
In conclusion, our study revealed that FSTL3, as a novel oncogene in NSCLC, was regulated by DSCAM-AS1 and miR-122-5p, and involved in cancer progression. Our work provides us with a further insight into the molecular mechanism of NSCLC, new molecular targets for its treatment and theoretical evidence for future studies.
Data Sharing Statement
The data used to support the findings of this study are available from the corresponding author upon request.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that they have no competing interest in this work.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi:10.3322/caac.21387
2. Allemani C, Weir HK, Carreira H, et al. Global surveillance of cancer survival 1995-2009: analysis of individual data for 25, 676, 887 patients from 279 population-based registries in 67 countries (CONCORD-2). Lancet. 2015;385:977–1010. doi:10.1016/S0140-6736(14)62038-9
3. Łochowski M, Łochowska B, Rębowski M, Brzeziński D, Cieślik-wolski B, Kozak J. Five-year survival analysis and prognostic factors in patients operated on for non-small cell lung cancer with N2 disease. J Thorac Dis. 2018;10(6):3180–3186. doi:10.21037/jtd
4. Kralisch S, Hoffmann A, Klöting N, et al. FSTL3 is increased in renal dysfunction. Nephrol Dial Transplant. 2017;32(10):1637–1644. doi:10.1093/ndt/gfw472
5. Mukherjee A, Sidis Y, Mahan A, et al. FSTL3 deletion reveals roles for TGF-beta family ligands in glucose and fat homeostasis in adults. Proc Natl Acad Sci U S A. 2007;104(4):1348–1353. doi:10.1073/pnas.0607966104
6. Oldknow KJ, Seebacher J, Goswami T, et al. Follistatin-like 3 (FSTL3) mediated silencing of transforming growth factor β (TGFβ) signaling is essential for testicular aging and regulating testis size. Endocrinology. 2013;154(3):1310–1320. doi:10.1210/en.2012-1886
7. Nam J, Perera P, Gordon R, et al. Follistatin-like 3 is a mediator of exercise-driven bone formation and strengthening. Bone. 2015;78:62–70. doi:10.1016/j.bone.2015.04.038
8. Liu G, Chen S, Deng S, et al. Association of follistatin-like 3 concentrations in serum and synovial fluid with the radiographic severity of knee osteoarthritis. Int J Clin Exp Med. 2015;8(10):18884–18888.
9. Razanajaona D, Joguet S, Ay AS, et al. Silencing of FLRG, an antagonist of activin, inhibits human breast tumor cell growth. Cancer Res. 2007;67(15):7223–7229. doi:10.1158/0008-5472.CAN-07-0805
10. Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10(3):155–159. doi:10.1038/nrg2521
11. Yang YR, Zang SZ, Zhong CL, et al. Increased expression of the IncRNA PVT1 promotes tumorigenesis in non-small cell lung cancer. Int J Clin Exp Pathol. 2014;7(10):6929.
12. Zhao W, Luo J, Jiao S. Comprehensive characterization of cancer subtype associated long non-coding RNAs and their clinical implications. Sci Rep. 2014;13(4):6591.
13. Liao J, Xie N. Long noncoding RNA DSCAM-AS1 functions as an oncogene in non-small cell lung cancer by targeting BCL11A. Eur Rev Med Pharmacol Sci. 2019;23(3):1087–1092. doi:10.26355/eurrev_201902_16998
14. Liu B, Qu J, Xu F, et al. MiR-195 suppresses non-small cell lung cancer by targeting CHEK1. Oncotarget. 2015;20:9445–9456.
15. Xu X, Gao F, Wang J, et al. MiR-122-5p inhibits cell migration and invasion in gastric cancer by down-regulating DUSP4. Cancer Biol Ther. 2018;19(5):427–435. doi:10.1080/15384047.2018.1423925
16. Li C, Qin F, Hu F, et al. Characterization and selective incorporation of small non-coding RNAs in non-small cell lung cancer extracellular vesicles. Cell Biosci. 2018;8:2. doi:10.1186/s13578-018-0202-x
17. Qin H, Sha J, Jiang C, et al. miR-122 inhibits metastasis and epithelial-mesenchymal transition of non-small-cell lung cancer cells. Onco Targets Ther. 2015;8:3175–3184. doi:10.2147/OTT.S91696
18. Wang L, Dong X, Ren Y, et al. Targeting EHMT2 reverses EGFR-TKI resistance in NSCLC by epigenetically regulating the PTEN/AKT signaling pathway. Cell Death Dis. 2018;9(2):129. doi:10.1038/s41419-017-0120-6
19. Wachters FM, Wong LS, Timens W, et al. ERCC1, hRad51, and BRCA1 protein expression in relation to tumour response and survival of stage III/IV NSCLC patients treated with chemotherapy. Lung Cancer. 2005;50(2):211–219. doi:10.1016/j.lungcan.2005.06.013
20. Ni X, Cao X, Wu Y, et al. FSTL1 suppresses tumor cell proliferation, invasion and survival in non-small cell lung cancer. Oncol Rep. 2018;39(1):13–20. doi:10.3892/or.2017.6061
21. Yang W, Wu Y, Wang C, et al. FSTL1 contributes to tumor progression via attenuating apoptosis in a AKT/GSK-3β - dependent manner in hepatocellular carcinoma. Cancer Biomark. 2017;20(1):75–85. doi:10.3233/CBM-170132
22. Zhang D, Ma X, Sun W, et al. Down-regulated FSTL5 promotes cell proliferation and survival by affecting Wnt/β-catenin signaling in hepatocellular carcinoma. Int J Clin Exp Pathol. 2015;8(3):3386–3394.
23. Zawadzka AM, Schilling B, Cusack MP, et al. Phosphoprotein secretome of tumor cells as a source of candidates for breast cancer biomarkers in plasma. Mol Cell Proteomics. 2014;13(4):1034–1049. doi:10.1074/mcp.M113.035485
24. Behnke M, Reimers M, Fisher R. The expression of embryonic liver development genes in hepatitis C induced cirrhosis and hepatocellular carcinoma. Cancers (Basel). 2012;4(3):945–968. doi:10.3390/cancers4030945
25. Bloise E, Couto HL, Massai L, et al. Differential expression of follistatin and FLRG in human breast proliferative disorders. BMC Cancer. 2009;9:320. doi:10.1186/1471-2407-9-320
26. Wang H, Tan Z, Hu H, et al. microRNA-21 promotes breast cancer proliferation and metastasis by targeting LZTFL1. BMC Cancer. 2019;19(1):738. doi:10.1186/s12885-019-5951-3
27. Gong ZH, Zhou F, Shi C, et al. miRNA-221 promotes cutaneous squamous cell carcinoma progression by targeting PTEN. Cell Mol Biol Lett. 2019;24:9. doi:10.1186/s11658-018-0131-z
28. Yin Z, Xu M, Li P. miRNA-221 acts as an oncogenic role by directly targeting TIMP2 in non-small-cell lung carcinoma. Gene. 2017;620:46–53. doi:10.1016/j.gene.2017.04.007
29. Tao Y, Lei L, Jing L, et al. MiRNA-10a is upregulated in NSCLC and may promote cancer by targeting PTEN. Oncotarget. 2015;6(30):30239–30250. doi:10.18632/oncotarget.4972
30. Sun C, Li S, Zhang F, et al. Long non-coding RNA NEAT1 promotes non-small cell lung cancer progression through regulation of miR-377-3p-E2F3 pathway. Oncotarget. 2016;7(32):51784–51814. doi:10.18632/oncotarget.10108
31. Yun C, Fuming Z, Chunkai Z, et al. Upregulated lncRNA SNHG1 contributes to progression of non-small cell lung cancer through inhibition of miR-101-3p and activation of Wnt/β-catenin signaling pathway. Oncotarget. 2017;8(11):17785–17794. doi:10.18632/oncotarget.14854
32. Liang WH, Li N, Yuan ZQ, et al. DSCAM-AS1 promotes tumor growth of breast cancer by reducing miR-204-5p and up-regulating RRM2. Mol Carcinog. 2019;58(4):461–473. doi:10.1002/mc.22941
33. Ji D, Hu G, Zhang X, et al. Long non-coding RNA DSCAM-AS1 accelerates the progression of hepatocellular carcinoma via sponging miR-338-3p. Am J Transl Res. 2019;11(7):4290–4302.
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.