Back to Journals » OncoTargets and Therapy » Volume 12
Up-regulation of CKAP2L expression promotes lung adenocarcinoma invasion and is associated with poor prognosis
Authors Xiong GS, Li LY, Chen XB, Song SN, Zhao YP, Cai WK, Peng JP
Received 1 August 2018
Accepted for publication 19 December 2018
Published 12 February 2019 Volume 2019:12 Pages 1171—1180
DOI https://doi.org/10.2147/OTT.S182242
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Leo Jen-Liang Su
Guosheng Xiong,1,* Liyin Li,2,* Xiaobo Chen,1 Sinuo Song,3 Yunping Zhao,1 Wenke Cai,4 Jingping Peng4
1Department of Thoracic Surgery, The First Affiliated Hospital of Kunming Medical University, Kunming, Yunnan 650032, People’s Republic of China; 2Department of Hematology, The First Affiliated Hospital of Kunming Medical University, Kunming, Yunnan 650032, People’s Republic of China; 3Department of Medical Management, Kunming General Hospital, Kunming, Yunnan 650032, People’s Republic of China; 4Department of Cardiovascular and Thoracic, Kunming General Hospital, Kunming, Yunnan 650032, People’s Republic of China
*These authors contributed equally to this work
Aim: The purpose of this study is to consider the function of cytoskeleton-associated protein 2-like (CKAP2L) in lung adenocarcinoma (LAD) development and its prognostic value.
Methods: The mRNA expression of CKAP2L and its correlation with clinical factors in LAD patients were analyzed from the data taken from The Cancer Genome Atlas and The First Affiliated Hospital of Kunming Medical University. We constructed H460 and A549 cell lines with silenced CKAP2L using RNA interference. Cell counting kit-8 assay and colony formation assays were carried out to determine the function of CKAP2L in H460 and A549 cell proliferation. Transwell and wound healing assays were applied to determine the effect of CKAP2L on H460 and A549 cell invasion and migration. The influences of CKAP2L on mitogen-activated protein kinase signaling pathway-related proteins were tested by Western blotting.
Results: CKAP2L expression is enhanced in LAD tissues and is predictive of poor prognosis in LAD patients. High expression of CKAP2L is associated with stage (P<0.001), lymph node status (P=0.002), and metastasis (P=0.025). Depletion of CKAP2L dramatically suppressed the proliferation, migration, and invasion of H460 and A549 cells. Moreover, the ratio of p-MEK/MEK and p-ERK/ERK reduced obviously in A549 cells after depleting CKAP2L.
Conclusion: Our findings implied that CKAP2L might be a promoter of LAD and could serve as a predictor for LAD patients.
Keywords: CKAP2L, prognosis, lung adenocarcinoma, migration, proliferation
Introduction
Cytoskeleton-associated protein 2-like (CKAP2L) gene is composed of 9 exons and encodes a protein that is considered to be indispensable to neural stem or progenitor cell division.1 This protein has been described as Radmis (radial fiber and mitotic spindle protein) based on its location to the radial fiber and mitotic spindle in neural stem/progenitor cells (NSPCs).1 Moreover, CKAP2L has been demonstrated to be a component of human centrosome, and is situated in the spindle, the midbody, and spindle pole.2 Current studies provide evidences proving that loss-of-function mutations in CKAP2L is one of the main cause of Filippi syndrome.3 A tight regulation of CKAP2L expression is very crucial for appropriate mitosis of neural precursor cells since overexpression of this protein leads to dramatic increase in cell mitosis.3 So far, the expression and function of CKAP2L in other tissues and in cancer progression are unknown.
Lung cancer is a leading cause of cancer-related death worldwide, as lung adenocarcinoma (LAD) is the most common histological type of it.4–7 Although much progress has been made in LAD therapy, its prognosis still remains poor.8 Furthermore, although advances in molecular-targeted therapy have improved treatment in patients with molecular mutations, it eventually fails owing to drug resistance.9,10 Hence, a deeper understanding of the molecular mechanism of LAD and efforts toward prophylaxis and early diagnosis will help in treating this disease.
Here, we analyzed the expression of CKAP2L in LAD tissues and the prognostic value of this gene in LAD. The function of CKAP2L in LAD cell proliferation and motility was investigated by constructing H460 and A549 cells with silenced CKAP2L. Finally, we quested the potential mechanism of how CKAP2L affects LAD cell proliferation and motility. Our data manifested that CKAP2L might be a facilitator in the development of LAD and could be used as a candidate predictor in LAD patients.
Methods
Data collection and patients
The mRNA expression data of 535 LAD tissues and 59 normal lung tissues were collected from The Cancer Genome Atlas (TCGA, https://tcga-data.nci.Nih.gov/tcga). In addition, 52 LAD cases resected at the First Affiliated Hospital of Kunming Medical University from 2008 to 2012 were gathered and analyzed in this paper. Informed consent was obtained from these 52 LAD patients. All the patients received no preoperative radiotherapy and/or chemotherapy. The clinical–pathological characteristics of these 52 samples are shown in Table 1.
  | Table 1 The clinical–pathological characteristics of the 52 lung adenocarcinoma patients |
Cell culture
Human LAD cell lines H460, A549, SPC-A1, and normal lung epithelial cell line BEAS-2B were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). H460 cells, SPC-A1 cells, and A549 cells were cultured in Roswell Park Memorial Institute (RPMI)-1640 medium and incubated at 37°C with 5% CO2. BEAS-2B cells were cultivated in DMEM/F12. FBS (10%), penicillin (100 U/mL), and streptomycin (100 mg/mL) were contained in the culture media.
Quantitative reverse transcription PCR (qRT-PCR)
Total RNA was extracted from the 52 LAD samples and the corresponding paracancer tissues as well as H460, A549, SPC-A1, and BEAS-2B cell lines using RNAiso Plus (TaKaRa Biotechnology, Dalian, China). Then cDNA were synthesized using a HiFiScript cDNA Synthesis Kit (CwBio, Beijing, China) with the extracted RNA as template. Afterwards, we utilized Applied Biosystems 7300 Sequence Detection System (Applied Biosystems, Foster City, CA, USA) to determine the mRNA expression level of CKAP2L. The primers used for qPCR were: CKAP2LF-5′- CAGGAGTACCTTGCAGCCAA-3′, CKAP2LR:5′- TGCTGATGGACAGATTTAGAAGGT-3′; GAPDHF: 5′-GGAGCGAGATCCCTCCAAAAT-3′, GAPDHR: 5′-GGCTGTTGTCATACTTCTCATGG-3′, where GAPDH was applied for normalization. Relative expression of CKAP2L was computed utilizing the 2−ΔΔCT method.11
Transfection
CKAP2L siRNA1 (si-CKAP2L1: 5′-AAACUGUCACUGAAGAGUC-3′), CKAP2L siRNA2 (si-CKAP2L2: 5′-ACUGUCACUGAAGAGUCAU-3′), and scrambled siRNA (si-con: 5′-GAAGUCACUGUCAAACUAG-3′) were synthesized by GENEWIZ Co., Ltd. (Suzhou, China). The siRNA was transfected into LAD cells utilizing Lipofectamine 2000 (Invitrogen, Shanghai, China). Afterwards, the cells were kept at 37°C for 6 hours followed by replacement of the media with fresh medium. After 48 hours of cultivation, cells were used to carry out the following tests.
Cell counting kit-8 (CCK8) test
After being transfected with si-CKAP2L or si-con, A549 and H460 cells were inoculated into a 96-well plate and cultured under standard conditions. The proliferation of these cells was determined using CCK8 at 24-, 48-, 72-, and 96-hour time points following the manufacturer instructions. The proliferation curves were plotted using GraphPad Prism 5.0.
Colony formation assay
Single cell suspensions of A549 and H460 cells with depleted CKAP2L were prepared and plated into 12-well plates (500 cells/well) with RPMI-1640 medium. After being conventionally cultured for 2 weeks, the cells were fixed with 4% paraformaldehyde and stained by 0.1% crystal violet dye. Finally, we counted the number of clones under light microscopy with 2× magnification.
Wound healing assay
A549 and H460 cells with depleted CKAP2L were plated on a 6-well plate and grown to confluence. Then, we made a horizontal wound using a 200-μL pipette tip after the cells had been starved for 24 hours. After that, we captured the wounds and measured their width under an optical microscope (Olympus, Tokyo, Japan) after 0 hour and 24 hours scratching. The assays were carried out for at least three times.
Transwell invasion and migration assays
The invasive and migration ability of A549 and H460 cells with silenced CKAP2L were analyzed utilizing Transwell assays. For invasion tests, the cells were seeded into the upper chambers, which were pre-coated with Matrigel (BD Biosciences, Franklin Lakes, NJ, USA) and then went into a 24 hours cultivation. Afterwards, we fixed the invasive cells with 4% paraformaldehyde and stained them using 0.1% crystal violet dye. The invasive cells were photographed in five random fields and counted under a light microscope with 200× magnification. The procedure of tranwell migration assay was similar to invasion assays. However, the upper chambers were not coated with Matrigel. All the experiments were performed in triplicate.
Western blot
We extracted total protein from A549 cells after silencing CKAP2L and performed SDS-PAGE to separate the proteins. The proteins were then electrotransferred to the polyvinylidene fluoride membranes (Millipore, Bedford, MA, USA) and blocked by 5% nonfat milk. Afterwards, the membranes were incubated with the primary anti-bodies (ERK, p-ERK, MEK, p-MEK, and GAPDH) and secondary anti-bodies successively. The expression of GAPDH was used as internal reference. Finally, the protein bands were detected using an enhanced chemiluminescence plus detection kit (Thermo Fisher Scientific, Inc., Waltham, MA, USA). All the experiments were performed three times.
Statistical analysis
Statistical analyses were performed using SPSS 15.0 software (IBM, Chicago, IL, USA) and the values were shown as mean ± SD. Student’s t-test was carried out to compare the difference between two groups. One-way ANOVA was utilized to compare the difference between multiple groups. Both the samples obtained from TCGA database and collected by ourselves were plotted into high CKAP2L expression and low CKAP2L expression groups on the basis of the median of CKAP2L mRNA expression. Kaplan–Meier analysis was used to analyze the relationship between CKAP2L expression and overall survival with log-rank test applied for comparison. Pearson’s chi-squared (χ2) test was performed to determine the association between CKAP2L expression and clinical–pathological factors. Cox proportional hazards model was performed to identify independent prognostic predictors. It was considered as significant when P<0.05.
Results
CKAP2L is overexpressed in LAD and is predictive of poor prognosis
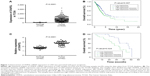
On the basis of the data taken from the TCGA database, we identified that the mRNA expression level of CKAP2L was dramatically higher in LAD tissues (n=535) than that in normal lung tissues (n=59) (Figure 1A, P<0.0001). Likely, by analyzing the mRNA expression of CKAP2L in the 52 LAD tissue samples that were collected in The First Affiliated Hospital of Kunming Medical University, we observed higher expression of CKAP2L in LAD tissues than in normal paracancer tissues (Figure 1C, P<0.0001). By Kaplan–Meier analysis, we discovered that higher CKAP2L expression was positively correlated with shorter overall survival both based on the TCGA database (Figure 1B, P=0.007) and the clinical data collected by ourselves (Figure 1D, P=0.024).
In correlation analysis (Table 2), statistically significant association was observed between CKAP2L expression and stage (P<0.001), lymph node status (P=0.002), or metastasis (P=0.025). No significant relationship was found between CKAP2L expression and age, gender, and T-stage. For the purpose of examining whether CKAP2L can be used as an independent predictor of prognosis in LAD, Cox regression analysis was carried out. By univariate analysis, we found that CKAP2L expression (P=0.0298), stage (P=0.03), T-stage (P=0.011), metastasis (P=0.005), and lymph node status (P=0.001) can be regarded as prognostic factors (Table 3). Multivariate analysis revealed that metastasis (P=0.015) and lymph node status (P=0.003) could be used as independent prognostic factors in LAD (Table 3).
  | Table 2 The association between CKAP2L expression and the clinical–pathological factors of 52 lung adenocarcinoma patients |
Knockdown of CKAP2L suppresses proliferation of LAD cells
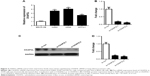
By qRT-PCR, we found that the expression of CKAP2L is dramatically higher in LAD cell lines H460, A549, and SPC-A1 than that in normal lung cell line BEAS-2B (Figure 2A, P<0.01). With the purpose of exploring the effect of CKAP2L expression on LAD cell growth, we used RNA interference to reduce the expression of CKAP2L gene in LAD cells and constructed LAD cells with silenced CKAP2L. As shown in Figure 2B–D, both the mRNA and protein expression levels of CKAP2L were dramatically reduced after transfected with si-CKAP2L 1 or si-CKAP2L 2 compared with the si-NC group. In the following experiments, we selected si-CKAP2L 2 to reduce CKAP2L expression in A549 and H460 cells. By CCK8 assays, we observed that depletion of CKAP2L remarkably decreased the OD value at 24, 48, and 72 hours in both A549 and H460 cells (P<0.05, Figure 3A–B). In colony formation assays, similar results were observed. The numbers of formatted A549 and H460 colonies in si-CKAP2L groups were dramatically less than that in the corresponding si-con groups (P<0.05, Figure 3C–D). These results demonstrated that down-regulation of CKAP2L exhibited an inhibitory effect on A549 and H460 cell proliferation, implying that CKAP2L promoted LAD progression.
Knockdown of CKAP2L suppresses migration and invasion of LAD cells
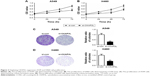
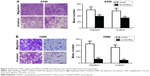
To investigate whether CKAP2L expression was involved in cell migration and invasion, Transwell and wound healing assays were carried out in A549 and H460 cells transfected with si-CKAP2L or si-con. As we predicted, knockdown of CKAP2L dramatically reduced the numbers of migrated and invasive A549 (Figure 4A) as well as H460 cells (Figure 4B) (P<0.01). Similarly, in wound healing assays, depletion of CKAP2L significantly reduced the migration distances of A549 cells (Figure 5A) as well as CKAP2L cells (Figure 5B). These observations implied that CKAP2L was a positive regulator of LAD motility.
Knockdown of CKAP2L suppresses mitogen-activated protein kinase (MAPK) signaling pathway
Afterwards, we explored the effect of knockdown of CKAP2L on MAPK signaling pathway in A549 cells by Western blot. Our results showed that the ratio of phospho-MAPK/extracellular signal-regulated kinase (ERK) kinase (MEK)/MEK and p-ERK/ERK were reduced obviously in A549 cells after depleting CKAP2L (Figure 6, P<0.01). This observation suggested that CKAP2L might act as a positive regulator in LAD progression by regulating MAPK signaling pathway.
Discussion
Out of control mitosis is a distinguishing characteristic of tumor cells, and this feature is effectively used to treat cancer by utilizing anti-tubulin drugs.12 However, high-grade LAD is often highly chemo-sensitive and these treatment is usually failed due to drug resistance. Therefore, it is of great significance to get insightful understanding into the mechanism of LAD progression and develop effective prophylaxis and early diagnosis methods. CKAP2L, also called radmis, is a microtubule-associated protein that appears at the mitotic-phase via the post-translational regulation and takes vital part in neural progenitor cell division.1 Down-regulation of this gene triggered the multipolar spindles and chromosome segregation in neural stem/progenitor cells (NSPCs).1 To date, little is known about the function of CKAP2L in cancer development. CKAP2 is an important paralog of CKAP2L. Previous reports have demonstrated that CKAP2 is up-regulated in primary human gastric adenocarcinomas. In breast cancer, CKAP2 was suggested to be an independent prognostic marker.13 In addition, the overexpression and oncogenic nature of CKAP2 has also been illustrated in prostate cancer,14 ovarian cancer,15 and glioma.16 Therefore, we supposed that CKAP2L might also participate in the occurrence and development of lung cancer.
In our present study, we identified that CKAP2L was overexpressed in LAD tissues (Figure 1A and C), and this overexpression is closely associated with poor prognosis of LAD patients (Figure 1B and D). Chi-squared test revealed that higher CKAP2L expression seems to be positively correlated with higher stage, lymph node status, and metastasis. Univariate analysis showed that stage, T-stage, metastasis and lymph node status, and CKAP2L expression could serve as prognostic factors for LAD. However, the result of multivariate analysis found that only metastasis (P=0.015) and lymph node status (P=0.003) could be considered as independent prognostic factors for overall survival although we observed a trend toward a correlation between high CKAP2L expression and poor outcomes (P=0.430). A larger sample size is needed to verify our current analysis results. The expression changes and prognostic value of CKAP2L implied that it probably plays an important promotional part in the occurrence and progression of LAD.
We then explored the influence of silencing CKAP2L on LAD cell proliferation and motility. The results showed that knockdown of CKAP2L dramatically impaired A549 and H460 cell proliferation, migration, and invasion abilities in vitro, indicating that CKAP2L functioned as a facilitator in LAD progression. In the migration and invasion assays, we observed that the H460 cells were more susceptible to CKAP2L silencing than A549 cells. This difference might be attributed to the discrimination between these two cell lines. H460 cells were isolated from pleural effusion of a patient with lung cancer. While A549 cell line was isolated from lung cancer tissues of a male. The expression level of p53 in H460 cells was similar to that in normal cells, and the keratin and vimentin fiber staining result was positive. As a novel mitotic spindle protein, we supposed that CKAP2L might affect LAD cell proliferation by participating in polymerization/stabilization of microtubules and then regulating cell mitosis.1 MAPK signaling pathway has been demonstrated to take part in diverse fundamental cellular processes, such as cell proliferation, migration, division, and death.17–19 Moreover, the involvement of this pathway in proliferation has been reported in various cancer types,20–22 including LAD.23 Since silencing CKAP2L exhibits similar inhibitory effect on H460 and A549 cell proliferation and motility, we only explored the effect of knockdown CKAP2L on MAPK signaling pathway in A549 cells. We observed that knockdown of CKAP2L dramatically reduced the ratio of p-MEK/MEK and p-ERK/ERK (P<0.01). This data suggested that CKAP2L might promote cell proliferation partially by regulating MAPK signaling pathway. However, further investigation is needed to find out how CKAP2L talks with the MAPK signaling pathway.
Conclusion
Our results strongly suggested that CKAP2L plays an important facilitating role in the development of LAD and is predictive of poor prognosis of LAD patients. In the future studies, we will carry out the experiments in animal models and attempt to clarify the possible mechanisms that underlie CKAP2L involvement in LAD proliferation and migration.
Acknowledgment
This study was supported by The Medical Discipline Backup Talent Cultivation Object in Yunnan province H-2017014.
Author contributions
All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
Yumoto T, Nakadate K, Nakamura Y, et al. Radmis, a novel mitotic spindle protein that functions in cell division of neural progenitors. PLoS One. 2013;8(11):e79895. | ||
Jakobsen L, Vanselow K, Skogs M, et al. Novel asymmetrically localizing components of human centrosomes identified by complementary proteomics methods. Embo J. 2011;30(8):1520–1535. | ||
Hussain MS, Battaglia A, Szczepanski S, et al. Mutations in CKAP2L, the human homolog of the mouse Radmis gene, cause Filippi syndrome. Am J Hum Genet. 2014;95(5):622–632. | ||
Collisson EA, Campbell JD, Brooks AN; Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511(7511):543–550. | ||
Liu J, Liu L, Cao L, Wen Q. Keratin 17 promotes lung adenocarcinoma progression by enhancing cell proliferation and invasion. Med Sci Monit. 2018;24:4782–4790. | ||
Wang X, He M, Li J, Wang H, Huang J. KLF15 suppresses cell growth and predicts prognosis in lung adenocarcinoma. Biomed Pharmacother. 2018;106:672–677. | ||
Qin H, Zhou J, Xu J, et al. The nuclear transcription factor RelB functions as an oncogene in human lung adenocarcinoma SPC-A1 cells. Cancer Cell Int. 2018;18(1):88. | ||
Reck M, Heigener DF, Mok T, Soria J-C, Rabe KF. Management of non-small-cell lung cancer: recent developments. The Lancet. 2013;382(9893):709–719. | ||
Katono K, Sato Y, Kobayashi M, et al. Clinicopathological significance of S100A14 expression in lung adenocarcinoma. Oncol Res Treat. 2017;40(10):594–602. | ||
Saito M, Shiraishi K, Kunitoh H, Takenoshita S, Yokota J, Kohno T. Gene aberrations for precision medicine against lung adenocarcinoma. Cancer Science. 2016;107(6):713–720. | ||
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25(4):402–408. | ||
Ahmed AA, Lu Z, Jennings NB, et al. SIK2 is a centrosome kinase required for bipolar mitotic spindle formation that provides a potential target for therapy in ovarian cancer. Cancer Cell. 2010;18(2):109–121. | ||
Sim SH, Bae CD, Kwon Y, et al. CKAP2 (cytoskeleton-associated protein2) is a new prognostic marker in HER2-negative luminal type breast cancer. PLoS One. 2017;12(8):e0182107. | ||
Yu G, Lee YC, Cheng CJ, et al. RSK promotes prostate cancer progression in bone through ING3, CKAP2, and PTK6-mediated cell survival. Mol Cancer Res. 2015;13(2):348–357. | ||
Gao Y, Liu X, Li T, et al. Cross-validation of genes potentially associated with overall survival and drug resistance in ovarian cancer. Oncol Rep. 2017;37(5):3084–3092. | ||
Wang K, Huang R, Li G, et al. CKAP2 expression is associated with glioma tumor growth and acts as a prognostic factor in high-grade glioma. Oncol Rep. 2018;40(4):2036–2046. | ||
Lai CS, Lee JH, Ct HO. Rosmanol potently inhibits lipopolysaccharide-induced iNOS and COX-2 expression through downregulating MAPK, NF-κB, STAT3 and C/EBP signaling pathways. J Agric Food Chem. 2011;3(3):198–206. | ||
Zhang P, Wu C, Huang XH, et al. Aspirin suppresses TNF-α-induced MMP-9 expression via NF-κB and MAPK signaling pathways in RAW264.7 cells. Exp Ther Med. 2017;14(6):5597–5604. | ||
Xu T, Lv Z, Chen Q, Guo M, Wang X, Huang F. Vascular endothelial growth factor over-expressed mesenchymal stem cells-conditioned media ameliorate palmitate-induced diabetic endothelial dysfunction through PI-3K/AKT/m-TOR/eNOS and p38/MAPK signaling pathway. Biomed Pharmacother. 2018;106:491–498. | ||
Chen DL, Hu ZQ, Zheng XF, et al. EDAG-1 promotes proliferation and invasion of human thyroid cancer cells by activating MAPK/ERK and Akt signal pathways. Cancer Biol Ther. 2016;17(4):414–421. | ||
Gong Y, He H, Liu H, Zhang C, Zhao W, Shao RG. Phosphorylation of myofibrillogenesis regulator-1 activates the MAPK signaling pathway and induces proliferation and migration in human breast cancer MCF7 cells. FEBS Lett. 2014;588(17):2903–2910. | ||
Liao T, Wen D, Ma B, et al. Yes-associated protein 1 promotes papillary thyroid cancer cell proliferation by activating the ERK/MAPK signaling pathway. Oncotarget. 2017;8(7):11719–11728. | ||
Zhou Q, Gui S, Zhou Q. Melatonin inhibits proliferation of human lung adenocarcinoma A549 cell line possibly through MAPK signal pathway. Acta Universitatis Medicinalis Anhui. 2014;9(7):e101132. |
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.