Back to Journals » Cancer Management and Research » Volume 13
Up-Regulation of circEIF6 Contributes to Pancreatic Cancer Development Through Targeting miR-557/SLC7A11/PI3K/AKT Signaling
Authors Zhang T, Li M, Lu H, Peng T
Received 3 September 2020
Accepted for publication 30 November 2020
Published 12 January 2021 Volume 2021:13 Pages 247—258
DOI https://doi.org/10.2147/CMAR.S280307
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Xueqiong Zhu
Tiequan Zhang, Mi Li, Haofeng Lu, Tao Peng
Department of Hepatobiliary Surgery, The First People’s Hospital of Jingzhou, Jingzhou 434000, People’s Republic of China
Correspondence: Tiequan Zhang
Department of Hepatobiliary Surgery, The First People’s Hospital of Jingzhou, No. 8 Hangkong Road, Shashi District, Jingzhou, Hubei 434000, People’s Republic of China
Tel +86-716-8115036
Email [email protected]
Background: Accruing evidences have pointed out that abnormal expression of circular RNAs (circRNAs) was closely related to the development of many malignancies. The present study intended to disclose the role of circRNA eukaryotic translation initiation factor 6 (circEIF6; hsa_circ_0060055) in pancreatic cancer progression.
Methods: Quantitative real-time polymerase chain reaction (qRT-PCR) was conducted to determine the expression of circEIF6, EIF6 messenger RNA (mRNA), microRNA-557 (miR-557) and solute carrier family 7 member 11 (SLC7A11) mRNA. Cell proliferation ability, migration and invasion abilities and apoptosis were evaluated by Cell Counting Kit 8 (CCK8) assay, transwell migration and invasion assays and flow cytometry. Western blot assay was performed for the expression determination of all proteins. The predicted interaction between miR-557 and circEIF6 or SLC7A11 was confirmed by dual-luciferase reporter assay. Xenograft tumor model was used for exploring the biological function of circEIF6 in vivo.
Results: CircEIF6 abundance was aberrantly up-regulated in pancreatic tumor tissues and cell lines. Cell proliferation, migration and invasion were significantly restrained while cell apoptosis was induced with the silencing of circEIF6 in pancreatic cancer cells. CircEIF6 silencing also hampered the activation of phosphatidylinositol 3-kinase (PI3K)/AKT serine/threonine kinase (AKT) pathway. CircEIF6 bound to miR-557, and circEIF6 silencing elevated the expression of miR-557 in pancreatic cancer cells. MiR-557 knockdown partly overturned circEIF6 silencing-induced effects in pancreatic cancer cells. SLC7A11 was a target of miR-557, and miR-557 overexpression suppressed malignant potential of pancreatic cancer cells partly through reducing the expression of SLC7A11. CircEIF6 knockdown blocked xenograft tumor growth in vivo.
Conclusion: CircEIF6 aggravated pancreatic cancer development through promoting cell proliferation, migration and invasion and suppressing cell apoptosis through targeting miR-557/SLC7A11/PI3K/AKT signaling.
Keywords: pancreatic cancer, EIF6, miR-557, SLC7A11, PI3K/AKT signaling
Introduction
Pancreatic cancer is a common digestive tract cancer that ranks the fourth in cancer-associated deaths globally.1,2 Most pancreatic cancer patients were diagnosed at an advanced condition at the first visit because of the asymptomatic onset of pancreatic cancer.3 Therefore, understanding the potential mechanism that drives pancreatic cancer development is important for developing effective diagnostic techniques and therapeutic strategies.
Circular RNAs (circRNAs) are characterized by stable circular structure due to the covalently closed ends.4 Recently, circRNAs have become the focus in the fields of human diseases and cancers.5,6 Accruing evidences have shown that circRNAs function as modulators in cancer development, and they may be novel bio-markers and therapeutic targets.4,7 CircRNA eukaryotic translation initiation factor 6 (circEIF6; hsa_circ_0060055) was abnormally up-regulated in pancreatic ductal adenocarcinoma tissues compared with adjacent normal tissues.8 Nevertheless, the precise contribution of circEIF6 in pancreatic cancer development is largely unknown.
MicroRNAs (miRNAs) are small non-coding RNAs that have been extensively studied in human cancer field.9 Many miRNAs serve as oncogenes or tumor suppressors in pancreatic cancer.10 For instance, miR-9-5p functioned as a tumor suppressor to suppress the malignant behaviors of pancreatic cancer cells through targeting GOT1.11 Zhang et al demonstrated that miR-135b-5p accelerated the aggressive phenotypes of pancreatic cancer cells through targeting NR3C2.12 MiR-557 overexpression restrained the proliferation capacity and invasion capacity in pancreatic cancer cells through targeting EGFR.13 Here, the mechanism of miR-557 in pancreatic cancer progression was further explored.
MiRNAs generally function as regulators in cancer development through targeting messenger RNAs (mRNAs).14 Solute carrier family 7 member 11 (SLC7A11) functions in modulating the transportation of amino acid.15,16 Former works have pointed out that the expression of SLC7A11 was up-regulated in cancers, and a high level of SLC7A11 was associated with malignant behaviors of cancer cells.17,18 Zhu et al claimed that miR-139-5p hampered cellular malignant proliferation and motility of pancreatic cancer cells through targeting SLC7A11/phosphatidylinositol 3-kinase (PI3K)/AKT serine/threonine kinase (AKT) signaling.19 Here, we investigated the potential mechanism behind the oncogenic role of SLC7A11 in pancreatic cancer.
The current study intended to analyze the expression pattern of circEIF6 in pancreatic cancer and investigate the downstream molecular mechanism by which circEIF6 contributes to pancreatic cancer progression.
Materials and Methods
Clinical Samples
Pancreatic tumor tissues (n=39) along with matched adjacent normal tissues (n=39; at least 2 cm away from the tumor border) were collected from pancreatic cancer patients at The First People’s Hospital of Jingzhou. No subject had received neoadjuvant treatment, and all subjects had signed informed consents before surgical resection. There were 17 patients with pancreatic tumors of I and II phase, while there were 22 patients with pancreatic tumors of III phase according to the Tumor-Node-Metastasis (TNM) clinical staging criteria.20 All protocols were given permission by the ethics committee of The First People’s Hospital of Jingzhou and was carried out according to the guidelines of the Declaration of Helsinki.
Cell Lines
Normal human pancreatic duct epithelial cell line (HPDE) and two pancreatic cancer cell lines (Hs 766T and SW1990) were purchased from BeNa Culture Collection (Beijing, People's Republic of China) and cultured with Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Carlsbad, CA, USA) added with 10% fetal bovine serum (FBS; Gibco) and 1% antibiotic (100 units/mL penicillin/100 μg/mL streptomycin; Solarbio, Beijing, People's Republic of China) in a 37°C incubator with 5% CO2/95% air.
Cyclization Validation
RNA samples extracted from Hs 766T or SW1990 cells were divided into two equal parts, and 2 μg RNA samples were incubated with 6U RNase R EIF6 and linearEIF6 mRNA were examined by qRT-PCR.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
RNA samples from tissues and cells were prepared with Trizol reagent (Invitrogen, Carlsbad, CA, USA). For miR-557 analysis, complementary DNA (cDNA) was synthesized using First-Strand cDNA Synthesis Kit (Takara, Dalian, People's Republic of China), and PCR was conducted using SYBR Premix Ex TaqTMEIF6, EIF6 mRNA and SLC7A11 mRNA, RNA was converted into cDNA with One Step TB GreenTM PrimeScriptTM RT-PCR Kit (Bio-Rad, Hercules, CA, USA) followed by PCR reaction using iQ SYBR Green Supermix (Bio-Rad). The specific primer sequences are shown in Table 1. Glyceraldehyde-3-EIF6, EIF6 mRNA and SLC7A11 mRNA, while U6 acted as the control for miR-557. The 2−ΔΔCt method was used for the quantification of these molecules.
 |
Table 1 Primers Used in qRT-PCR |
Cell Transfection
CircEIF6 small interfering RNAs (siRNAs), including si-circEIF6#1 (5ʹ-UCGAACACACCGGAGGGAACU-3ʹ), si-circEIF6#2 (5ʹ-AACACACCGGAGGGAACUGCA-3ʹ) and si-circEIF6#3 (5ʹ-ACACACCGGAGGGAACUGCAC-3ʹ), negative control siRNA (si-NC), circEIF6 short hairpin RNA (sh-circEIF6), sh-NC, miR-557 mimics (miR-557), miR-NC, miR-557 inhibitor (in-miR-557) and in-miR-NC were purchased from GenePharma (Shanghai, People's Republic of China). For the overexpression of SLC7A11, we introduced the full length of SLC7A11 into pcDNA vector to obtain SLC7A11 recombinant overexpression plasmid, and pcDNA empty vector was used as the control. Lipofectamine 3000 reagent (Invitrogen) was used to transfect these RNA and plasmid into Hs 766T or SW1990 cells.
Cell Counting Kit 8 (CCK8) Assay
Hs 766T or SW1990 cells were digested and seeded into 96-well plates. After a fixed time interval after transfection, pancreatic cancer cells were incubated with CCK8 reagent (20 μL; Dojindo Laboratories, Kumamoto, Japan) for 1 hour. The absorbance at 450 nm was measured by the microplate reader (Nikon, Tokyo, Japan).
Transwell Migration and Invasion Assays
A total of 40 μL Matrigel (BD biosciences, San Jose, CA, USA) at the dilution of 1:8 was added to the transwell upper chambers for 30 minutes at 37°C for solidification to conduct transwell invasion assay, while un-coated transwell chambers were used for transwell migration assay. After transfection for 24 hours, pancreatic cancer cells in serum-free medium were seeded into the upside of the membrane, while the lower chambers were filled with medium added with 10% FBS. Transwell plates were placed in a cell incubator for 24 hours. Cells migrated or invaded to the lower surface of the membrane were washed with phosphate buffer saline (PBS, Sangon Biotech, Shanghai, People's Republic of China), fixed with 4% paraformaldehyde (Sigma, St. Louis, MO, USA) and stained with 0.5% crystal violet (Sangon Biotech). The numbers of migrated and invaded cells in five random fields were counted at 100 × magnification. The scale bar indicates 200 μM.
Flow Cytometry
After transfection for 72 hours, the DNA content and phosphatidylserine in transfected Hs 766T or SW1990 cells were stained using propidium iodide (PI; R&D systems, Abingdon, UK) and fluorescein isothiocyanate (FITC)-labeled Annexin V (Annexin V-FITC; R&D systems) for 15 minutes in the dark, respectively. The proportion of apoptotic cells (the first quadrant and the fourth quadrant) was analyzed by FC-500 flow cytometer (Beckman Coulter, Pasadena, CA, USA).
Western Blot Assay
After transfection for 24 hours, Hs 766T or SW1990 cells were lysed by Western cell lysis buffer (Beyotime, Haimen, People's Republic of China). An equal amount of protein samples was loaded onto sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gel and blotted onto polyvinylidene fluoride (PVDF) membrane (Millipore, Billerica, MA, USA). The membrane was incubated with 5% skimmed milk for 1 hour at room temperature for blocking. Primary antibodies against p-AKT (07–1398; Sigma), AKT (SAB4500797; Sigma), p-PI3K (ab389562; Abcam, Cambridge, MA, USA), PI3K (ab32089; Abcam), SLC7A11 (SAB2500951; Sigma) and β-actin (A1978; Abcam) were incubated with the membrane at 4°C overnight. Afterwards, horseradish-peroxidase (HRP)-labeled secondary antibody (Sigma) was incubated with the membrane for 2 hours at room temperature. Immunochemical detection was conducted with the enhanced chemiluminescence (ECL) Detection System (GE Healthcare, Chicago, IL, USA).
Bioinformatic Prediction
Circular RNA Interactome software (https://circinteractome.nia.nih.gov) was used to predict circEIF6-miRNA interactions, while the mRNA targets of miR-557 were predicted by TargetScan software (http://www.targetscan.org).
Dual-Luciferase Reporter Assay
CircEIF6 or SLC7A11 3ʹ untranslated region (3ʹUTR) sequence, containing the wild-type (WT) miR-557 binding sites, was fused to the luciferase reporter vector pmirGLO vector (Promega, Madison, WI, USA), termed as circEIF6 WT and SLC7A11 3ʹUTR WT. Also, site-directed mutagenesis in miR-557-binding sites in circEIF6 or SLC7A11 3ʹUTR sequence was made to generate circEIF6 MUT and SLC7A11 3ʹUTR MUT. Hs 766T or SW1990 cells were co-transfected with miR-NC or miR-557 and luciferase plasmids with Lipofectamine 3000 reagent (Invitrogen), and the luciferase activity was determined by the Dual-Luciferase Reporter System Kit (Promega) after transfection for 48 hours.
Xenograft Tumor Model
Animal experiments were conducted with the permission of the animal care and use committee of The First People’s Hospital of Jingzhou and performed in accordance with the guidelines of the National Animal Care and Ethics Institution. SW1990 cell line stably transfected with sh-EIF6 or sh-NC was established. Afterwards, SW1990 cells stably transfected with sh-EIF6 or sh-NC were subcutaneously inoculated into the right flank of BALB/c male nude mice (Vital River Laboratory Animal Technology, Beijing, People's Republic of China) in sh-NC group and sh-EIF6 group. The volume of tumors was recorded every week using the caliper as width2 × length × 0.5. After inoculation for 28 days, these nude mice were euthanized and the tumors were dissected and weighed. The levels of circEIF6 and miR-557 were detected by qRT-PCR, while Western blot assay was conducted to examine the levels of SLC7A11, p-AKT, AKT, p-PI3K and PI3K.
Statistical Analysis
The statistical results are represented as mean ± standard deviation. Each experiment was repeated for three times. Statistical analysis was conducted by Student’s t-test (in two groups) or one-way analysis of variance (ANOVA) followed by Tukey’s test (in more than two groups) using GraphPad Prism 7.0 software. Linear correlation among circEIF6, miR-557 and SLC7A11 was assessed by Pearson correlation coefficient. Difference was defined as statistically significant with the P value of less than 0.05.
Results
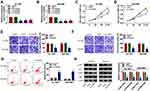
CircEIF6 is Highly Expressed in Pancreatic Cancer Tissues and Cell Lines
CircEIF6 expression was significantly enhanced in pancreatic tumor tissues relative to adjacent normal tissues (Figure 1A). Pancreatic cancer patients were grouped into I+II group (n=17) and III group (n=22) on the basis of TNM staging. As displayed in Figure 1B, circEIF6 expression was observed to be notably enhanced in tumor tissues of advanced stage (III stage) relative to that in I+II stage, suggesting that circEIF6 expression was positively related to the malignant grade of pancreatic cancer. ROC curve was generated to assess the diagnostic value of circEIF6 expression in tumor tissues of pancreatic cancer patients. The ROC curve revealed an area under the curve (AUC) of 0.9093 (95% confidence interval (CI)=0.845–0.9735) (Figure 1C). Furthermore, a significant elevation in circEIF6 expression was discovered in two pancreatic cancer cell lines relative to HPDE cell line (Figure 1D). RNase R was used to evaluate the circular characteristics of circEIF6. RNA samples from Hs 766T and SW1990 cells were treated with RNase R or not, and we found that the level of linearEIF6 rather than circEIF6 was dramatically reduced upon RNase R digestion (Figure 1E and F), providing strong evidence that circEIF6 possessed circular structure. Overall, abnormal expression of circEIF6 might imply its important function in pancreatic cancer progression.
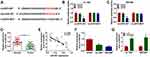
CircEIF6 Knockdown Hampers Cell Proliferation, Migration, Invasion and Induces Cell Apoptosis of Pancreatic Cancer Cells
To characterize the function of circEIF6 in pancreatic cancer, we designed three siRNAs targeting circEIF6 to silence its expression in pancreatic cancer cells. As shown in Figure 2A and B, among three siRNAs of circEIF6, si-circEIF6#1 (written as si-EIF6 in the following experiments) was selected for the following experiments due to it’s the highest transfection efficiency. As examined by CCK8 assay, cell proliferation was significantly suppressed with the silencing of circEIF6 compared with si-NC-transfected pancreatic cancer cells (Figure 2C and D). As mentioned in Figure 2E and F, cell migration ability and invasion ability were both significantly restrained following the transfection of si-EIF6 compared with si-NC group. As determined by flow cytometry, cell apoptosis was triggered following the transfection of si-EIF6 than that in si-NC group (Figure 2G). The activity of PI3K/AKT signaling was analyzed through measuring the levels of p-AKT, AKT, p-PI3K and PI3K in pancreatic cancer cells. As displayed in Figure 2H, after silencing circEIF6, the levels of p-AKT/AKT and p-PI3K/PI3K were both reduced than that in si-NC group in the two pancreatic cancer cell lines. Taken together, circEIF6 silencing blocked cell proliferation, migration, invasion and promoted cell apoptosis of pancreatic cancer cells.
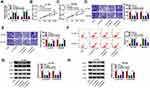
MiR-557 is a Target of circEIF6 in Pancreatic Cancer Cells
Bioinformatic search using Circular RNA Interactome was conducted to identify the possible miRNA targets of circEIF6. Among all the candidate miRNA targets (Circular RNA Interactome software) of circEIF6, only miR-557 has been reported to be down-regulated in pancreatic cancer,13 thus we performed dual-luciferase reporter assay to test the target interaction between miR-557 and circEIF6. The complementary sequence between circEIF6 and miR-557 is shown in Figure 3A. Dual-luciferase reporter assay was performed in pancreatic cancer cells to further verify the target interaction between miR-557 and circEIF6. Transfection with miR-557 rather than miR-NC significantly reduced the luciferase intensity of luciferase reporter circEIF6 WT in Hs 766T and SW1990 cells (Figure 3B and C). Furthermore, luciferase activities were unchanged with the transfection of reporter circEIF6 MUT upon the co-transfection of miR-NC or miR-557 (Figure 3B and C), providing evidence that miR-557 was a target of circEIF6 in pancreatic cancer cells. MiR-557 expression was significantly decreased in pancreatic tumor tissues when compared with adjacent normal tissues (Figure 3D). An obvious negative correlation was observed between the expression of circEIF6 and miR-557 in pancreatic tumor tissues (Figure 3E). Compared with HPDE cell line, miR-557 expression was notably reduced in Hs 766T and SW1990 cells (Figure 3F). CircEIF6 silencing caused a significant up-regulation in miR-557 expression in both the two pancreatic cancer cell lines (Figure 3G). Taken together, circEIF6 interacted with miR-557 in pancreatic cancer cells.
CircEIF6 Knockdown Restrains the Malignant Phenotypes of Pancreatic Cancer Cells Through Enhancing miR-557 Level
To explore if circEIF6-induced influences in pancreatic cancer cells required the involvement of miR-557, we co-transfected Hs 766T and SW1990 cells with si-NC, si-circEIF6, si-EIF6 + in-miR-NC or si-EIF6 + in-miR-557. As displayed in Figure 4A, circEIF6 knockdown-mediated up-regulation of miR-557 level was largely mitigated by the addition of miR-557 inhibitor (in-miR-557). CircEIF6 interference suppressed cell proliferation as evidenced by the cell proliferation curve shown in Figure 4B and C, and miR-557 silencing partly rescued the proliferation ability of pancreatic cancer cells. The migration and invasion abilities were restrained by the silencing of circEIF6, while these effects were largely attenuated by the introduction of in-miR-557 (Figure 4D and E). CircEIF6 interference triggered the apoptosis of pancreatic cancer cells, which was suppressed by the addition of in-miR-557 (Figure 4F). The levels of p-AKT/AKT and p-PI3K/PI3K were reduced with the knockdown of circEIF6, while these influences were largely counteracted by the silencing of miR-557 (Figure 4G and H), suggesting that circEIF6 interference suppressed the activity of PI3K/AKT pathway partly through up-regulating miR-557. These results together demonstrated that miR-557 silencing attenuated circEIF6 knockdown-mediated effects in the biological behaviors of pancreatic cancer cells.
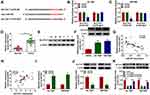
SLC7A11 is a Target of miR-557 in Pancreatic Cancer Cells
Bioinformatic software (TargetScan) was used to investigate miR-557-mRNAs interactions. Among all the predicted mRNA targets of miR-557, we screened five mRNA targets that have been reported to be up-regulated in pancreatic cancer, including HMGB2,21 SOX2,22 FOXM1,23 MACC124 and SLC7A1119 (Supplementary Figure 1A). The regulatory relationships between miR-557 and these five candidate targets in pancreatic cancer cells were tested by qRT-PCR. As shown in Supplementary Figure 1B and 1C, miR-557 overexpression reduced the expression of all these five candidate mRNA targets, and SLC7A11 was selected for the following experiments due to its the highest obvious negative regulatory relationship with miR-557. The predicted complementary sites between SLC7A11 3ʹUTR and miR-557 are shown in Figure 5A. Three bases “CAA” in the complementary sites of SLC7A11 were mutated by “GUU” to test if SLC7A11interacted with miR-557 via this predicted sequence. Transfection of miR-557 significantly reduced the luciferase activity with the co-transfection of SLC7A11 3ʹUTR WT reporter plasmid compared with miR-NC and SLC7A11 3ʹUTR WT group (Figure 5B and C). Moreover, transfection with miR-557 or miR-NC in SLC7A11 3ʹUTR MUT group caused indiscriminate luciferase activity (Figure 5B and C), suggesting that SLC7A11 interacted with miR-557 via “UGCAAA” sequence. The expression characteristics of SLC7A11 were subsequently explored. SLC7A11 mRNA and protein expression was enhanced in pancreatic tumor tissues compared with that in adjacent normal tissues (Figure 5D and E). The level of SLC7A11 protein was notably enhanced in two pancreatic cancer cell lines relative to that in HPDE cell line (Figure 5F). Linear correlation between the expression of SLC7A11 and miR-557 or circEIF6 was analyzed by Pearson correlation coefficient. As displayed in Figure 5G and H, SLC7A11 expression was negatively correlated with the expression of miR-557, while there was a positive correlation between the expression of SLC7A11 and circEIF6. The overexpression efficiency of miR-557 was assessed by qRT-PCR. The expression of miR-557 was significantly enhanced in pancreatic cancer cells following transfection with miR-557 mimics rather than miR-NC (Figure 5I). After overexpressing miR-557, SLC7A11 expression was significantly decreased in both Hs 766T and SW1990 cells (Figure 5J). Hs 766T and SW1990 cells were transfected with si-EIF6 alone or together with in-miR-557 to explore the regulatory relationship between circEIF6 and SLC7A11. As mentioned in Figure 5K, circEIF6 silencing reduced the protein expression of SLC7A11, while this suppressive effect induced by circEIF6 knockdown was largely alleviated by the addition of in-miR-557. Taken together, circEIF6 silencing reduced SLC7A11 expression through enhancing miR-557 level in pancreatic cancer cells.
SLC7A11 Overexpression Partly Alleviates miR-557-Mediated Effects in Pancreatic Cancer Cells
Hs 766T and SW1990 cells were transfected with miR-557 alone or together with SLC7A11 plasmid to conduct rescue experiments to explore if miR-557-mediated effects were based on its negative regulatory relationship with SLC7A11. MiR-557 overexpression down-regulated the expression of SLC7A11, while the level of SLC7A11 was largely rescued with the addition of SLC7A11 overexpression plasmid (Figure 6A). MiR-557 overexpression resulted in significant suppression in the abilities of proliferation, migration and invasion in pancreatic cancer cells, while these suppressive effects were all attenuated by the addition of SLC7A11 plasmid (Figure 6B–E). Cell apoptosis was triggered with the forced expression of miR-557, and the co-transfection of miR-557 and SLC7A11 alleviated the apoptosis of pancreatic cancer cells (Figure 6F). MiR-557 overexpression down-regulated the phosphorylation levels of AKT and PI3K, while the addition of SLC7A11 plasmid largely recovered the levels of p-AKT/AKT and p-PI3K/PI3K in pancreatic cancer cells (Figure 6G and H). Overall, miR-557 suppressed the malignant potential and the activity of PI3K/AKT signaling through targeting SLC7A11.
CircEIF6 Interference Suppresses the Growth of Pancreatic Cancer Xenograft Tumors in vivo
Given the pro-tumor role of circEIF6 in pancreatic cancer in vitro, we subsequently explored the function of circEIF6 in tumor growth in vivo. SW1990 cells stably expressing sh-EIF6 or sh-NC were inoculated into the nude mice. The volume of tumors from sh-EIF6-expressing cells was suppressed compared to those from the sh-NC group (Figure 7A). Also, tumor weight was notably reduced with the interference of circEIF6 compared with the sh-NC group (Figure 7B). The expression of circEIF6 and the protein level of SLC7A11 were reduced in tumor tissues from sh-EIF6-expressing SW1990 cells compared to those from the sh-NC-expressing SW1990 cells (Figure 7C and E). An opposite tendency in miR-557 expression was found in tumor tissues when compared with the expression trend of circEIF6 or SLC7A11 (Figure 7D). The levels of p-AKT/AKT and p-PI3K/PI3K were both reduced in the sh-EIF6 group compared with that in the sh-NC group (Figure 7F). Overall, circEIF6 silencing suppressed pancreatic cancer progression in vivo.
Discussion
The discovery of circRNAs provides a novel insight into cancer treatment, which is of vital significance for identifying new diagnostic and therapeutic targets for cancers.25,26 Many circRNAs were dysregulated in diverse malignancies, and the clinical significance of dysregulated circRNAs in cancers has been investigated by former works. For instance, circ-LDLRAD3 was highly expressed in the serum samples and tissue samples of pancreatic cancer patients, and high level of circ-LDLRAD3 was related to aggressive phenotype in pancreatic cancer patients.27 Bi et al claimed that circ-ZKSCAN1 expression was reduced in bladder cancer tissues and cell lines, and low expression of circ-ZKSCAN1 was closely associated with the malignant characteristics in bladder cancer patients.28 CircEIF6 exhibited higher expression in pancreatic ductal adenocarcinoma tissue specimens than that in adjacent normal tissue specimens.8 However, the biological significance behind the up-regulation of circEIF6 in pancreatic cancer remains to be uncovered. In line with a previous article,8 we found that circEIF6 expression was elevated in pancreatic tumor tissues and cell lines. Furthermore, patients with pancreatic cancer in III phase had higher expression of circEIF6 compared with that in I+II phase. CircEIF6 silencing hampered the proliferation, migration and invasion and promoted the apoptosis of pancreatic cancer cells. PI3K/AKT signaling played important roles in suppressing cell apoptosis and inducing cell proliferation, and abnormal activation of this signaling has been proved to accelerate the progression of many malignancies.29,30 For instance, circ_001569 was proved to be an unfavorable prognostic factor to accelerate the proliferation and motility of breast cancer cells through activating PI3K/AKT signaling.31 The forced expression of circ_0000520 attenuated the herceptin resistance of gastric cancer cells through suppressing PI3K/AKT signaling.32 We found that circEIF6 interference also restrained the activity of PI3K/AKT signaling in pancreatic cancer cells.
Through performing bioinformatic analysis and dual-luciferase reporter assay, miR-557 was verified as a target of circEIF6 in pancreatic cancer cells. MiR-557 has been reported to restrain the development of pancreatic cancer and lung cancer. For example, Yang et al demonstrated that miR-557 overexpression suppressed cell proliferation ability and invasion ability in pancreatic cancer cells through targeting EGFR.13 Qiu et al claimed that miR-557 suppressed lung cancer development through reducing LEF1 levels.33 MiR-557 expression was reduced in pancreatic tumor tissues and cell lines, and it was reversely modulated by circEIF6 in pancreatic cancer cells. When we simultaneously transfected si-EIF6 and in-miR-557 into pancreatic cancer cells, the tumor suppressive functions caused by circEIF6 silencing were largely counteracted by the interference of miR-557. These results demonstrated that circEIF6 exerted its functions partly based on its regulatory relationship with miR-557.
As predicted by TargetScan and verified by dual-luciferase reporter assay, SLC7A11 was confirmed as one of the targets of miR-557. SLC7A11 was reported to regulate the extracellular cysteine transportation.15,17 Accumulating evidence has been uncovered that aberrant expression of SLC7A11 is related to the preneoplastic lesions and the development of many malignancies.34,35 A high level of SLC7A11 was observed in many malignancies, and high abundance of SLC7A11 was associated with dismal prognosis and chemoresistance of cancer cells.36,37 For instance, Wu et al claimed that SLC7A11 expression was enhanced in oral squamous cell carcinoma tissues and cell lines, and the forced expression of miR-375 suppressed cell proliferation and invasion of oral squamous cell carcinoma cells through suppressing SLC7A11.38 Zhu et al demonstrated that miR-139-5p hampered the proliferation and motility of pancreatic cancer cells through suppressing SLC7A11, thus inactivating PI3K/AKT signaling.19 SLC7A11 was observed to be up-regulated in pancreatic tumor tissues and cell lines. Furthermore, circEIF6 silencing reduced the expression of SLC7A11 through elevating miR-557 expression in pancreatic cancer cells. MiR-557 overexpression restrained the proliferation, migration and invasion and facilitated the apoptosis of pancreatic cancer cells, while SLC7A11 overexpression largely regained these malignant behaviors in miR-557-overexpressed pancreatic cancer cells. CircEIF6 silencing also prominently hampered the growth of pancreatic cancer xenograft tumors in vivo.
In conclusion, circEIF6 accelerated the proliferation, migration, invasion and restrained the apoptosis of pancreatic cancer cells through targeting miR-557/SLC7A11/PI3K/AKT signal axis. CircEIF6/miR-557/SLC7A11/PI3K/AKT signaling might provide novel therapeutic targets for pancreatic cancer therapy.
Funding
There is no funding to report.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Kamisawa T, Wood LD, Itoi T, Takaori K. Pancreatic cancer. Lancet. 2016;388(10039):73–85. doi:10.1016/S0140-6736(16)00141-0
2. Marchesi F, Piemonti L, Mantovani A, Allavena P. Molecular mechanisms of perineural invasion, a forgotten pathway of dissemination and metastasis. Cytokine Growth Factor Rev. 2010;21(1):77–82. doi:10.1016/j.cytogfr.2009.11.001
3. McGuigan A, Kelly P, Turkington RC, Jones C, Coleman HG, McCain RS. Pancreatic cancer: A review of clinical diagnosis, epidemiology, treatment and outcomes. World J Gastroenterol. 2018;24(43):4846–4861. doi:10.3748/wjg.v24.i43.4846
4. Meng S, Zhou H, Feng Z, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16(1):94.
5. Gao JL, Chen G, He HQ, Wang J. [CircRNA as a new field in human disease research]. Zhongguo Zhong Yao Za Zhi. 2018;43(3):457–462.
6. Kristensen LS, Hansen TB, Venø MT, Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37(5):555–565.
7. Zhang HD, Jiang LH, Sun DW, Hou JC, Ji ZL. CircRNA: a novel type of biomarker for cancer. Breast Cancer. 2018;25(1):1–7.
8. Li H, Hao X, Wang H, et al. Circular RNA expression profile of pancreatic ductal adenocarcinoma revealed by microarray. Cell Physiol Biochem. 2016;40(6):1334–1344.
9. Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17(5):272–283.
10. Qadir MI, Faheem A. miRNA: A diagnostic and therapeutic tool for pancreatic cancer. Crit Rev Eukaryot Gene Expr. 2017;27(3):197–204. doi:10.1615/CritRevEukaryotGeneExpr.2017019494
11. Wang J, Wang B, Ren H, Chen W. miR-9-5p inhibits pancreatic cancer cell proliferation, invasion and glutamine metabolism by targeting GOT1. Biochem Biophys Res Commun. 2019;509(1):241–248. doi:10.1016/j.bbrc.2018.12.114
12. Zhang Z, Che X, Yang N, et al. miR-135b-5p Promotes migration, invasion and EMT of pancreatic cancer cells by targeting NR3C2. Biomed Pharmacother. 2017;96:1341–1348. doi:10.1016/j.biopha.2017.11.074
13. Yang Y, Sun KK, Shen XJ, Wu XY, Li DC. miR-557 inhibits the proliferation and invasion of pancreatic cancer cells by targeting EGFR. Int J Clin Exp Pathol. 2019;12(4):1333–1341.
14. Lee YS, Dutta A. MicroRNAs in cancer. Annu Rev Pathol. 2009;4(1):199–227. doi:10.1146/annurev.pathol.4.110807.092222
15. Sato H, Tamba M, Ishii T, Bannai S. Cloning and expression of a plasma membrane cystine/glutamate exchange transporter composed of two distinct proteins. J Biol Chem. 1999;274(17):11455–11458. doi:10.1074/jbc.274.17.11455
16. Bannai S. Transport of cystine and cysteine in mammalian cells. Biochim Biophys Acta. 1984;779(3):289–306. doi:10.1016/0304-4157(84)90014-5
17. Okuno S, Sato H, Kuriyama-Matsumura K, et al. Role of cystine transport in intracellular glutathione level and cisplatin resistance in human ovarian cancer cell lines. Br J Cancer. 2003;88(6):951–956. doi:10.1038/sj.bjc.6600786
18. Liu -X-X, Li X-J, Zhang B, et al. MicroRNA-26b is underexpressed in human breast cancer and induces cell apoptosis by targeting SLC7A11. FEBS Lett. 2011;585(9):1363–1367. doi:10.1016/j.febslet.2011.04.018
19. Zhu J-H, De Mello RA, Yan Q-L, et al. MiR-139-5p/SLC7A11 inhibits the proliferation, invasion and metastasis of pancreatic carcinoma via PI3K/Akt signaling pathway. Biochim Biophys Acta Mol Basis Dis. 2020;1866(6):165747. doi:10.1016/j.bbadis.2020.165747
20. Shida D, Kanemitsu Y, Hamaguchi T, Shimada Y. Introducing the eighth edition of the tumor-node-metastasis classification as relevant to colorectal cancer, anal cancer and appendiceal cancer: a comparison study with the seventh edition of the tumor-node-metastasis and the Japanese Classification of Colorectal, Appendiceal, and Anal Carcinoma. Jpn J Clin Oncol. 2019;49(4):321–328.
21. Cai X, Ding H, Liu Y, et al. Expression of HMGB2 indicates worse survival of patients and is required for the maintenance of Warburg effect in pancreatic cancer. Acta Biochim Biophys Sin (Shanghai). 2017;49(2):119–127.
22. Jiang J, Li Z, Yu C, et al. MiR-1181 inhibits stem cell-like phenotypes and suppresses SOX2 and STAT3 in human pancreatic cancer. Cancer Lett. 2015;356(2):962–970. doi:10.1016/j.canlet.2014.11.007
23. Jingyang Z, Jinhui C, Lu X, et al. Mir-320b inhibits pancreatic cancer cell proliferation by targeting FOXM1. Curr Pharm Biotechnol. 2020;27:100–109.
24. Wang G, Kang M-X, Lu W-J, et al. MACC1: A potential molecule associated with pancreatic cancer metastasis and chemoresistance. Oncol Lett. 2012;4(4):783–791. doi:10.3892/ol.2012.784
25. Patop IL, Kadener S. circRNAs in cancer. Curr Opin Genet Dev. 2018;48:121–127. doi:10.1016/j.gde.2017.11.007
26. Vo JN, Cieslik M, Zhang Y, et al. The landscape of circular RNA in cancer. Cell. 2019;176(4):869–881. doi:10.1016/j.cell.2018.12.021
27. Yang F, Liu D-Y, Guo J-T, et al. Circular RNA circ-LDLRAD3 as a biomarker in diagnosis of pancreatic cancer. World J Gastroenterol. 2017;23(47):8345–8354. doi:10.3748/wjg.v23.i47.8345
28. Bi J, Liu H, Dong W, et al. Circular RNA circ-ZKSCAN1 inhibits bladder cancer progression through miR-1178-3p/p21 axis and acts as a prognostic factor of recurrence. Mol Cancer. 2019;18(1):133. doi:10.1186/s12943-019-1060-9
29. Burris HA. Overcoming acquired resistance to anticancer therapy: focus on the PI3K/AKT/mTOR pathway. Cancer Chemother Pharmacol. 2013;71(4):829–842. doi:10.1007/s00280-012-2043-3
30. Hassan B, Akcakanat A, Holder AM, Meric-Bernstam F. Targeting the PI3-kinase/Akt/mTOR signaling pathway. Surg Oncol Clin N Am. 2013;22(4):641–664. doi:10.1016/j.soc.2013.06.008
31. Xu J-H, Wang Y, Xu D. Hsa_circ_001569 is an unfavorable prognostic factor and promotes cell proliferation and metastasis by modulating PI3K-AKT pathway in breast cancer. Cancer Biomarkers. 2019;25(2):193–201. doi:10.3233/CBM-182293
32. Lv X, Li P, Wang J, et al. hsa_circ_0000520 influences herceptin resistance in gastric cancer cells through PI3K-Akt signaling pathway. J Clin Lab Anal. 2020;23:e23449.
33. Qiu J, Hao Y, Huang S, et al. MiR-557 works as a tumor suppressor in human lung cancers by negatively regulating LEF1 expression. Tumour Biol. 2017;39(6):1010428317709467.
34. Wada T, Ishimoto T, Seishima R, et al. Functional role of CD44v-xCT system in the development of spasmolytic polypeptide-expressing metaplasia. Cancer Sci. 2013;104(10):1323–1329.
35. Drayton RM, Dudziec E, Peter S, et al. Reduced expression of miRNA-27a modulates cisplatin resistance in bladder cancer by targeting the cystine/glutamate exchanger SLC7A11. Clin Cancer Res. 2014;20(7):1990–2000.
36. Huang Y, Dai Z, Barbacioru C, Sadée W. Cystine-glutamate transporter SLC7A11 in cancer chemosensitivity and chemoresistance. Cancer Res. 2005;65(16):7446–7454.
37. Savaskan NE, Heckel A, Hahnen E, et al. Small interfering RNA-mediated xCT silencing in gliomas inhibits neurodegeneration and alleviates brain edema. Nat Med. 2008;14(6):629–632.
38. Wu Y, Sun X, Song B, Qiu X, Zhao J. MiR- 375/SLC7A11 axis regulates oral squamous cell carcinoma proliferation and invasion. Cancer Med. 2017;6(7):1686–1697.
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.