Back to Journals » Lung Cancer: Targets and Therapy » Volume 9
Uncommon EGFR mutations in cytological specimens of 1,874 newly diagnosed Indonesian lung cancer patients
Authors Syahruddin E , Wulandari L , Sri Muktiati N, Rima A, Soeroso N, Ermayanti S, Levi M, Hidajat H, Widjajahakim G, Utomo ARH
Received 14 October 2017
Accepted for publication 27 December 2017
Published 23 March 2018 Volume 2018:9 Pages 25—34
DOI https://doi.org/10.2147/LCTT.S154116
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Sai-Hong Ignatius Ou
Elisna Syahruddin,1,2 Laksmi Wulandari,3 Nunuk Sri Muktiati,4 Ana Rima,5 Noni Soeroso,6 Sabrina Ermayanti,7 Michael Levi,8 Heriawaty Hidajat,2 Grace Widjajahakim,8 Ahmad Rusdan Handoyo Utomo9
1Department of Pulmonology and Respiratory Medicine, Faculty of Medicine, Universitas Indonesia; 2Department of Pulmonology, Persahabatan Hospital, Jakarta, Indonesia; 3Department of Pulmonology and Respiratory Medicine, Faculty of Medicine, Universitas Airlangga – Soetomo Hospital, Surabaya, Indonesia; 4Department of Pulmonology and Respiratory Medicine, Faculty of Medicine, Universitas Brawijaya – Saiful Anwar General Hospital, Malang, Indonesia; 5Department of Pulmonology and Respiratory Medicine, Faculty of Medicine, Universitas Sebelas Maret, Dr. Moewardi General Hospital, Surakarta, Indonesia; 6Department of Pulmonology and Respiratory Medicine, Faculty of Medicine, University of Sumatera Utara, Adam Malik General Hospital, Medan, Indonesia; 7Department of Pulmonology and Respiratory Medicine, Faculty of Medicine, Andalas University, M. Djamil Hospital, Padang, Indonesia; 8Kalbe Genomics Laboratory, Division of Molecular Pathology Testing Service, PT Bifarma Adiluhung, 9Stem Cell and Cancer Institute, Cancer Diagnostic Research Division, PT Kalbe Farma Tbk, Jakarta, Indonesia
Purpose: We aimed to evaluate the distribution of individual epidermal growth factor receptor (EGFR) mutation subtypes found in routine cytological specimens.
Patients and methods: A retrospective audit was performed on EGFR testing results of 1,874 consecutive cytological samples of newly diagnosed or treatment-naïve Indonesian lung cancer patients (years 2015–2016). Testing was performed by ISO15189 accredited central laboratory.
Results: Overall test failure rate was 5.1%, with the highest failure (7.1%) observed in pleural effusion and lowest (1.6%) in needle aspiration samples. EGFR mutation frequency was 44.4%. Tyrosine kinase inhibitor (TKI)-sensitive common EGFR mutations (ins/dels exon 19, L858R) and uncommon mutations (G719X, T790M, L861Q) contributed 57.1% and 29%, respectively. Approximately 13.9% of mutation-positive patients carried a mixture of common and uncommon mutations. Women had higher EGFR mutation rate (52.9%) vs men (39.1%; p<0.05). In contrast, uncommon mutations conferring either TKI responsive (G719X, L861Q) or TKI resistance (T790M, exon 20 insertions) were consistently more frequent in men than in women (67.3% vs 32.7% or 69.4% vs 30.6%; p<0.05). Up to 10% EGFR mutation–positive patients had baseline single mutation T790M, exon 20 insertion, or in coexistence with TKI-sensitive mutations. Up to 9% patients had complex or multiple EGFR mutations, whereby 48.7% patients harbored TKI-resistant mutations. One patient presented third-generation TKI-resistant mutation L792F simultaneously with T790M.
Conclusion: Routine diagnostic cytological techniques yielded similar success rate to detect EGFR mutations. Uncommon EGFR mutations were frequent events in Indonesian lung cancer patients.
Keywords: EGFR mutations, lung cancer, treatment naive, T790M, tyrosine kinase inhibitor, Indonesia, cytology
Introduction
Lung cancer in Indonesia ranks the fourth most common of all cancers.1 Majority of lung cancer cases are found in late stages and cytological specimens are common sources of diagnostic practices in tertiary hospitals.1 In addition to valuable diagnostic tools, cytological specimens are useful sources of epidermal growth factor receptor (EGFR) mutation testing. Specific guidelines of EGFR mutation testing in cytological specimens have been issued and adopted widely.2,3 However, there are considerable concerns regarding EGFR testing failure rates that may delay timeline of treatment decisions. Few or lack of tumor cells, improper fixation procedures, poor extracted DNA quality, and/or absence of or generation of nonspecific polymerase chain reaction (PCR) products have led to testing failures.3
Indonesian health authority has published national formulary to reimburse expenses of tyrosine kinase inhibitors (TKI) as first-line treatment for lung cancer patients bearing EGFR mutation. In 2014, we had described common EGFR mutations associated with first-generation TKI (erlotinib and gefitinib) sensitivity mainly in exons 19 (insertions/deletions) and 21 (L858R) obtained from cytological specimens using Sanger sequencing.4 However, the prevalence and clinical pathology associations of rare or uncommon EGFR mutations such as G719S/A/C (collectively G719X), T790M, and L861Q had not been described extensively in Indonesia. These uncommon mutations are sensitive to second and third generations of EGFR TKI, namely afatinib and osimertinib.5 Specifically, T790M mutation rate is thought to be low in treatment-naïve patients, but it contributes up to 50% of patients who are resistant to first-generation TKI.6–8
In this real-world EGFR mutation testing of treatment-naïve lung cancer patients, we had employed combination of PCR high-resolution melt (HRM) and restriction fragment length polymorphism (RFLP) to screen for common EGFR mutations (exon 19 insertions/deletions and L858R mutation in exon 21 of EGFR gene) to improve EGFR genotyping sensitivity.9,10 PCR HRM allowed rapid screening for genetic mutations due to differential melting properties of wild-type and mutant alleles PCR products.11 Both PCR HRM and RFLP methods have demonstrated superior sensitivity to Sanger sequencing.12 We also described the impact of various cytological sampling techniques to successful testing rate and evaluated frequency of individual mutation subtypes as well as their clinical pathology associations.
Patients and methods
Patients
Since the initial introduction of EGFR testing a few years ago, Indonesian clinicians and pathologists had been routinely using cytological specimens as primary testing sources as practical approach. Such routine practices cited successful EGFR testing from cytological samples by a reputable Southeast Asian laboratory.13 Moreover, tissue resection or surgical biopsies were not commonly performed by most clinicians (personal communications). Consequently, there were no formalin-fixed paraffin-embedded (FFPE) tissue specimens being received by our EGFR testing facility. Cytological specimens of 1,874 consecutive newly diagnosed lung cancer patients were received and tested for EGFR mutation by Kalbe Genomics Laboratory from September 2015 to April 2016. Cytological specimens along with pathology reports describing sex, age, cytopathology, and diagnostic sampling methods were received from 44 cities in Indonesia. However, fixative procedures of cytological specimens were not described in the EGFR testing request forms, except for 175 samples that arrived as FFPE blocks.
Kalbe Genomics is an ISO15189 accredited laboratory for EGFR mutation testing and has demonstrated consistent satisfactory performance in EGFR proficiency testing organized by European Molecular Genetic Quality Network and UK NEQAS annually since 2011. Ethic committees of Faculty of Medicine Universitas Airlangga, Soetomo General Hospital, Surabaya, and Persahabatan Hospital, Jakarta, approved this study. The study was performed in accordance with the 1964 Helsinki Declaration and its later amendements. Patient identities were anonymized. The approving ethics committees waived the need for informed consent because the study was based on existing administrative records and clinical data.
EGFR mutation screening program in Indonesia
During the study period, Astra Zeneca Indonesia (AZI) and Roche Indonesia (RI) invited physicians to test for EGFR mutation in newly diagnosed lung cancer patients and covered the costs for any patients diagnosed with lung adenocarcinoma or any non-small-lung cancers, respectively. The program stipulated no obligation in any kind of prescription in compliance with both AZI and RI ethical code of conducts. Test results were sent directly from the laboratory to the physicians.
DNA extraction
Genomic DNA was isolated from tumors using the QIAamp DNA Micro kit (Qiagen, Hilden, Germany) following the manufacturer’s protocol. DNA from each sample was eluted in 50 µL of AE buffer (included in the kit).
Mutation analysis
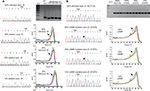
EGFR exons 19 and 21 mutation screenings were performed using PCR HRM analysis. PCR cycling and HRM analyses were performed on Rotor-Gene Q (Qiagen) using intercalating dye SYTO9 (Thermo Fisher Scientific Inc., Waltham, MA, USA) as described.9 Samples were denatured with an initial hold of 95°C for 30 s and a melting profile from 79°C to 90°C rising at 0.2°C. HRM data were presented as derivative graph to observed “split peak” indicating presence of mutated alleles (Figure 1) using Rotor-Gene Q software (Qiagen). Split peak pattern was observable in low percentage of mutant alleles, that is, less than 25%, a percentage that was usually not detectable using direct sequencing method. PCR RFLP method was then used to follow up suspected mutation suggested by presence of “split peak” pattern. Genotyping of EGFR L858R and L861Q hotspot mutations in exon 21 was performed using PCR RFLP that had been shown to detect 1% mutant allele.10
Mutations in EGFR exons 18 (namely to detect G719X) and 20 (T790M and insertions) were analyzed using Sanger sequencing. Primer pairs for EGFR exon 18 were designed with primer-BLAST software and for EGFR exon 20 was described previously.14 Some samples carrying mutation in exon 20 T790M were retested using Therascreen EGFR RGQ PCR kit (Qiagen, Manchester, UK) and/or AmoyDx EGFR 29 mutations detection kit (AmoyDx, Xiamen, China), according to the manufacturer’s instructions.
Direct sequencing
PCR amplification products were purified using the ExoSAP-IT PCR Product Cleanup (Affymetrix/USB, Santa Clara, CA, USA) according to the manufacturer’s protocol. Sequencing analysis was performed on an Applied Biosystem 3500 Genetic Analyzer. Nucleotide sequences of primers are available upon requests.
Statistical analysis
Fisher’s exact test was used to analyze associations between the presence of EGFR mutations and clinical pathology characteristics. Significance was set at p<0.05 (two-sided).
Results
Utility of melting peak PCR HRM to screen EGFR mutations
Direct sequencing method generally could not detect the presence of mutations when there were less than 25% as shown in left panels of Figure 1A (exon 19 mutations) and 1B (exon 21 mutations). To improve our capability to detect EGFR mutations, we employed PCR HRM to screen for the presence of EGFR mutations in exons 19 (Figure 1A right panel) and 21 (Figure 1B right panel) and observed different forms of melt pattern between PCR amplicons containing normal/wild-type EGFR and mutant EGFR samples when presented in derivative graph mode (Figure 1). As shown in Figure 1A, melt pattern (pointed by arrow) of wild-type exon 19 PCR amplicon (green trace line) showed single peak, while samples (red and blue trace lines) with PCR amplicons bearing mutational exon 19 insertions or deletions (ins/dels) demonstrated split peaks.
Similar split peak patterns were also observed in L858R sample (orange trace line, Figure 1B), while normal or wild-type samples showed single peak (green and brown trace lines). Unlike exon 19, however, split peak patterns shown in samples having mutations in exon 21 were due to single base substitution instead of multi PCR amplicons. Agarose electrophoreses of exon 21 PCR amplicon also confirmed the presence of single PCR product amplicon (Figure 1B upper panel). Therefore, split peak pattern strongly suggested the presence of mutant alleles.
To determine the analytical sensitivity of HRM approach, we tested using serial dilution of artificial DNA method. As shown in right panels of Figure 1A and 1B, threshold of detection limit indicated by the presence of split peak corresponded to 12% and 6% of mutant alleles in exons 19 and 21, respectively.
Impact of cytological techniques to EGFR testing
Clinical pathology characteristics of consecutive 1,874 newly diagnosed lung cancer patients drawn from 44 Indonesian cities are described in Table 1.
  | Table 1 Demography and clinical pathology characteristics (N=1,874 patients) |
Male patients (61%) were more frequent than female patients (39%). Median age was 57 years with a range between 19 and 92 years.
Most cytological specimens were adenocarcinoma (94%), obtained as malignant pleural effusion (MPE, 26%), as well as from fine needle aspirations (FNA, 20%), bronchoscopies (17%), and transthoracic needle biopsies (20%).
Out of the 1,874 consecutive samples, 95 (5.1%) samples failed EGFR testing. Failures were divided into preanalytical and analytical failures. Seventy-four (3.9%) samples were rejected outright (preanalytical failure), because the numbers of tumor cells were too few (<100 cells) or absent altogether.
MPE specimens showed the highest preanalytical failure rate (6.6%) and FNA specimens demonstrated the least (1.6%) (Table 2). Upon passing preanalytical step, there were only 11 samples that failed to generate specific amplicons after repeated PCR attempts at least twice. Out of the 11 samples, 7 were formalin-fixed while 4 were direct smear samples. Therefore, formalin fixation had higher frequency of PCR failure (2.4%; 4 out of 164 FFPE samples) than direct smear preparations (0.4% or 7 out of 1,626 samples; p=0.0019).
EGFR mutation frequency and clinical pathology associations
Overall EGFR mutation frequency was 44.5% (95% CI: 42.09–46.71). Approximately 57.1% and 29% of EGFR mutation–positive patients had common TKI-sensitive mutations (exon 19 ins/dels and L858R) and uncommon mutations (G719X, T790M, exon 20 insertions, and L861Q), respectively. The remaining 29% of patients harbored mixture of common and uncommon EGFR mutations (G719X, T790M, and L861Q) (Table 3).
  | Table 3 Breakdown of EGFR mutation types and rates Abbreviations: EGFR, epidermal growth factor receptor; TKI, tyrosine kinase inhibitor. |
Most patients harbored single mutations (80.5%). However, 19.5% of patients had multiple or complex mutations involving more than one mutation subtypes. Furthermore, first-generation TKI-resistant T790M mutations were found as single (3.4%) and complex (4.2%) TKI-sensitive mutations. The proportion of T790M in complex mutations (48.7%) was higher than in single mutations (9.6%, Table 3). Moreover, complex mutation cases of T790M/L858R (30%) were found more frequently than T790M/exon 19 ins/del (9%).
When stratified according to gender, EGFR mutations were higher in women (52.9%) than in men (39.1%, p<0.05). Furthermore, adenocarcinoma patients had higher rate of EGFR mutations (45.1%) than nonadenocarcinoma (34.3%, p=0.028) (Table 4).
  | Table 4 Associations between clinicopathology and types of EGFR mutations Abbreviations: NSCLC, non small cell lung carcinoma; EGFR, epidermal growth factor receptor; TKI, tyrosine kinase inhibitor. |
Common mutations (exon 19 ins/dels, L858R) conferring sensitivity to TKI were more prevalent in female (54.9%) than in male patients (45.1%). In contrast, uncommon EGFR mutations conferring either sensitivity (G719X, L861Q) or resistance to TKI (T790M, exon 20 insertions) were more frequent in male (64.9%) than in female patients (35.1%) (Table 4).
EGFR T790M mutation
Using Sanger sequencing, we found that majority of T790M mutations were heterozygous as shown in a typical sequencing result (Figure 2A upper panel), which we confirmed using real-time PCR (Figure 2B). Examination of T790M-positive cases revealed that up to 16% showed mutant allele specific imbalances (MASI) due to overrepresentation of mutant alleles (Figure 2A lower panel). To detect probable presence of germline mutation, DNA was isolated from adjacent normal cells. In 1out of 5 randomly selected MASI cases, a heterozygous T790M mutation was detected (Figure 2C) in normal cells indicating germline mutation. Confirmation in peripheral blood was not done because the patient died before the initiation of this study. In the other 4 cases, T790M mutations were absent in normal cells supporting the relatively rare frequency of T790M germline mutation. Last, we found that 1 out of 136 patients had concomitant T790M and L792F mutations, a putative resistance marker to second- and third-generation TKI (Figure 2D).
Discussion
We reported that the overall failure rate of EGFR mutation testing in real-world population was 5.1%, with MPE showing the highest (7.6%) and FNA the lowest (1.6%). MPE failure rates were combination of rejection during preanalytical step (6.6%) and PCR failures (1%) during analytical step. In addition, improper formalin fixation protocol performed at the referring hospitals to our laboratory might contribute to PCR failure. For instance, fixation time may affect the integrity of pre-PCR DNA template.15 However, an FFPE specimen failure rate of 2.4% obtained in our cohort was still lower than that of 11.4% obtained by the recent RING diagnostic trial involving 13 laboratories.16 Taken as a whole, our experience in receiving cytological specimens had similar success rate (94.9%) to previous descriptive review analyzing 19 publications of EGFR mutation testing in cytology samples stating an overall success rate of ~95%.3
The rate of total EGFR mutation in our population (44.5%) using cytological specimens was also similar to what laboratories in neighboring Southeast Asian countries had reported using either cytological13 or tissue specimens.17,18 Notably, the EGFR mutation rate was higher than our previous study (29%) when Sanger sequencing was used.4 The current study used PCR HRM and RFLP that had higher analytical sensitivity than Sanger sequencing and covered 4 exons (18–21) instead of just 2 exons (19 and 21). In addition, others and we had described the utility of split peak pattern of PCR HRM previously to screen for RAS and EGFR mutations rapidly.19,20
Most EGFR mutations in our current cohort were also detected as single mutations and the remaining 19.5% was complex mutations (containing more than one mutation). Our complex mutation rate was slightly higher than that demonstrated by other studies varying between 7% and 14%.21–24 Using massive parallel sequencing, the complex mutation rate is increased to 26%25 and has been associated with poor prognosis.
Most EGFR mutation studies to date reported major tyrosine kinase sensitizing mutations such as exon 19 insertions/deletions and L858R substitution mutations in exon 21. These common mutations generally comprised 80%–90% of total EGFR mutations, while the remaining mutations or uncommon mutations contributed 10%–20%.26,27 However, our population had high rates of uncommon mutations (composed of G719X, exon 20 insertions, T790M, and L861Q) contributing up to 43% of total EGFR mutations. High proportion of uncommon EGFR mutations to total EGFR mutations has been reported in European population (50%)28 and Chinese regions of Yunnan Province (70%).29
The reasons of variations in EGFR mutation subtypes may be attributed to geographical differences, ethnic backgrounds,30 and environmental exposures (coal burning, wooden smoke, cigarette smoking).29,31 Although information about smoking history was not available within our cohort, Indonesian lung cancer smoking attributable fractions in males is as high as 87% and in females 12%.32 This is consistent with the high prevalence of smoking among Indonesian males (65%), which ranks third in the world.33 Our data showed that male patients (65%; p<0.05) had higher rate of EGFR uncommon mutations than female patients (35%), which may be partly explained by recent descriptive studies suggesting putative association between uncommon mutations (G719X and L861Q) and smoking history.34,35 Therefore, future studies are needed to clarify definite association between EGFR uncommon mutations and smoking history in Indonesian lung cancer patients.
Among uncommon EGFR mutation types, T790M mutation has generated diagnostic as well as clinical interests. Up to 50% of common EGFR mutant patients would develop resistance to first-generation TKI due to acquired T790M mutations. TKI-naïve patients are not expected to harbor T790M mutations as confirmed by insensitive detection method such as Sanger sequencing with frequency typically less than 5%.6 Using Sanger we did find significant portion of T790M mutations in treatment-naïve patients (7.6%) having slightly higher frequency than what other studies have reported.8,36 T790M prevalence in complex mutation contributed up to 48.7% and mostly coexisted with L858R that is consistent with recent meta-analysis study of baseline T790M.37
Although Sanger sequencing is not as sensitive as amplification-refractory mutation system PCR, it may yield certain advantages. We were able to discern the extent of MASI by comparing the relative sequencing tracer heights of mutant allele vis a vis normal allele.38 We found that the rate of MASI in T790M mutations was similar to other mutations such as L858R and exon 19 insertions/deletions (26%–37%).39,40
Clinical significance of baseline T790M with or without MASI was not clear. However, presence of baseline T790M has been correlated with good prognosis,41 while others have demonstrated shorten progression free survival (PFS)42 and median overall survival.8 Nevertheless, current clinical practice does not exclude prescription of first-generation TKI to patients harboring concurrent T790M and TKI sensitive mutations.6 Interestingly, recent clinical trial subjecting 60 EGFR mutated patients (5 of whom harbored T790M mutations) to osimertinib as first-line treatment demonstrated an objective response rate of 77%43 and a PFS of 19.3 months, a significant extension of historical PFS of 10–13 months.44 Therefore, population with significant numbers of baseline T790M mutations like ours may benefit from using osimertinib as first-line treatment.
Furthermore, we had explored the potential presence of T790M germline mutations in some specimens. Out of the 5 specimens showing T790M MASI, we found 1 specimen having heterozygous T790M mutation in normal cells from the same slide. Unfortunately, we were not able to confirm mutation in the blood because the patient died at the time of our study. To our knowledge, this was the first study to demonstrate the utility of cytological specimens to screen for potential germline mutation. Germline mutation T790M has been an interest due to associated risk to develop lung cancer in family members having inherited the identical mutant alleles. Due to relatively rare mutation of T790M, the extent of germline T790M mutation may be as high as 50% of patients with somatic mutations.45 However, in general population, prevalence of germline T790M mutation has been estimated to be 1 in 7,500.46 We were not able to compare the proportion of germline T790M within our cohort, because majority of cytological slides whose tumor cells had been scraped during routine EGFR mutation testing were not available. Lastly, we also found one specimen having L792F mutation in complex with T790M mutation. L792F mutation has been proposed as putative resistant marker to second-generation (afatinib)47 and third-generation (osimertinib) TKI.48 L792F-acquired mutations had been shown in plasma of 3 patients who were resistant to osimertinib48 but not in pretreatment samples. Therefore, we found evidence that resistance marker to third-generation TKI may exist prior treatment albeit with extremely low frequency.
Conclusion
We used PCR HRM “split peak” melt pattern to screen and analyze EGFR mutation in real-world testing of Indonesian lung cancer samples obtained from major cities using routine cytological specimens. We found high rates of uncommon EGFR mutations (G719X, L861Q) in Indonesian male lung cancer patients, potential germline T790M mutation, and L792F next-generation TKI resistance EGFR mutation in cytological samples of untreated patients. These patients may benefit from first-line treatment using second- and third-generation TKIs.
Acknowledgments
We thank senior pulmonologists Drs Sita Laksmi, Jamal Zaini, and Achmad Hudoyo for reviewing patients’ clinical information and expert anatomic pathologists Winiarti Gani, Ruth Sembiring, and Patricia Diana for cytopathological confirmation of smears specimens. We appreciate Fitria Yunida, Siska Yogiwanti, dan Dewi Nawangwulan for excellent technical assistance. We also thank Najmiatul Masykura, Asep Ridwanulloh, Muhammad Yunus, and Audi Tri Harsono for early works validating rapid EGFR mutation screening methods and Cynthia Christina for organizing EGFR mutation database. Portion of EGFR mutation testing costs was provided by Astrazeneca Indonesia (AZI) and Roche Indonesia (RI). Testing results of individual patients remained confidential and were not accessible to AZI and RI.
Disclosure
The authors report no conflicts of interest in this work.
References
Prasetiyo PD, Pawitra I, Wijaya I. Expression of TTF-1 and CK-7 in the diagnosis of pleural effusion cytology suspected lung adenocarcinoma. J Biomed Transl Res. 2015;1(1):22–26. | ||
Roy-Chowdhuri S, Aisner DL, Allen TC, et al. Biomarker testing in lung carcinoma cytology specimens: a perspective from members of the Pulmonary Pathology Society. Arch Pathol Lab Med. April 2016:arpa.2016-0091-SA-7. | ||
da Cunha Santos G, Saieg MA. Preanalytic parameters in epidermal growth factor receptor mutation testing for non-small cell lung carcinoma: a review of cytologic series. Cancer Cytopathol. 2015;123(11):633–643. | ||
Yatabe Y, Kerr KM, Utomo A, et al. EGFR mutation testing practices within the Asia Pacific region: results of a multicenter diagnostic survey. J Thorac Oncol. 2015;10(3):438–445. | ||
Banno E, Togashi Y, Nakamura Y, et al. Sensitivities to various epidermal growth factor receptor-tyrosine kinase inhibitors of uncommon epidermal growth factor receptor mutations L861Q and S768I: what is the optimal epidermal growth factor receptor-tyrosine kinase inhibitor? Cancer Sci. 2016;107(8):1134–1140. | ||
Denis MG, Vallée A, Théoleyre S. EGFR T790M resistance mutation in non-small cell lung carcinoma. Clin Chim Acta. 2015;444:81–85. | ||
Yu HA, Arcila ME, Rekhtman N, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res. 2013;19(8):2240–2247. | ||
Yu HA, Arcila ME, Hellmann MD, Kris MG, Ladanyi M, Riely GJ. Poor response to erlotinib in patients with tumors containing baseline EGFR T790M mutations found by routine clinical molecular testing. Ann Oncol. 2014;25(2):423–428. | ||
Do H, Krypuy M, Mitchell PL, Fox SB, Dobrovic A. High resolution melting analysis for rapid and sensitive EGFR and KRAS mutation detection in formalin fixed paraffin embedded biopsies. BMC Cancer. 2008;8(1):142. | ||
Kawada I, Soejima K, Watanabe H, et al. An alternative method for screening EGFR mutation using RFLP in non-small cell lung cancer patients. J Thorac Oncol. 2008;3(10):1096–1103. | ||
Erali M, Voelkerding KV, Wittwer CT. High resolution melting applications for clinical laboratory medicine. Exp Mol Pathol. 2008;85(1):50–58. | ||
Ellison G, Zhu G, Moulis A, Dearden S, Speake G, McCormack R. EGFR mutation testing in lung cancer: a review of available methods and their use for analysis of tumour tissue and cytology samples. J Clin Pathol. 2013;66(2):79–89. | ||
Pang B, Matthias D, Ong CW, et al. The positive impact of cytological specimens for EGFR mutation testing in non-small cell lung cancer: a single South East Asian laboratory’s analysis of 670 cases. Cytopathology. 2012;23(4):229–236. | ||
Amann J, Kalyankrishna S, Massion PP, et al. Aberrant epidermal growth factor receptor signaling and enhanced sensitivity to EGFR inhibitors in lung cancer. Cancer Res. 2005;65(1):226–235. | ||
Cree IA, Deans Z, Ligtenberg MJL, et al. Guidance for laboratories performing molecular pathology for cancer patients. J Clin Pathol. 2014;67(11):1–11. | ||
Kapp JR, Diss T, Spicer J, et al. Variation in pre-PCR processing of FFPE samples leads to discrepancies in BRAF and EGFR mutation detection: a diagnostic RING trial. J Clin Pathol. 2015;68(2):111–118. | ||
Shi Y, Au JS-K, Thongprasert S, et al. A prospective, molecular epidemiology study of EGFR mutations in Asian patients with advanced non-small cell lung cancer of adenocarcinoma histology (PIONEER). J Thorac Oncol. 2014;9(2):154–162. | ||
Liam CK, Wong CK, Tan JL. EGFR mutation detection by polymerase chain reaction-direct sequencing and allele-specific real-time PCR. J Thorac Oncol. 2014;9(9):e71–e72. | ||
Levi M, Prayogi G, Sastranagara F, et al. Clinicopathological associations of K-RAS and N-RAS mutations in Indonesian colorectal cancer cohort. J Gastrointest Cancer. Epub 2017 Jan 3. | ||
Borràs E, Jurado I, Hernan I, et al. Clinical pharmacogenomic testing of KRAS, BRAF and EGFR mutations by high resolution melting analysis and ultra-deep pyrosequencing. BMC Cancer. 2011;11(1):406. | ||
Keam B, Kim D-W, Park JH, et al. Rare and complex mutations of epidermal growth factor receptor, and efficacy of tyrosine kinase inhibitor in patients with non-small cell lung cancer. Int J Clin Oncol. 2014;19(4):594–600. | ||
Hata A, Yoshioka H, Fujita S, Kunimasa K, Kaji R. Complex mutations in the epidermal growth factor receptor gene in non-small cell lung cancer. J Thorac Oncol. 2010;5(10):1524–1528. | ||
Hsieh M-H, Fang Y-F, Chang W-C, et al. Complex mutation patterns of epidermal growth factor receptor gene associated with variable responses to gefitinib treatment in patients with non-small cell lung cancer. Lung Cancer. 2006;53(3):311–322. | ||
Beau-Faller M, Prim N, Ruppert AM, et al. Rare EGFR exon 18 and exon 20 mutations in non-small cell lung cancer on 10 117 patients: a multicentre observational study by the French ERMETIC-IFCT network. Ann Oncol. 2013;25(1):126–131. | ||
Kim EY, Cho EN, Park HS, et al. Compound EGFR mutation is frequently detected with co-mutations of actionable genes and associated with poor clinical outcome in lung adenocarcinoma. Cancer Biol Ther. 2015;17(3):237–245. | ||
Castellanos E, Feld E, Horn L. Driven by mutations: the predictive value of mutation subtype in EGFR-mutated non-small cell lung cancer. J Thorac Oncol. 2017;12(4):612–623. | ||
Kuiper JL, Hashemi SMS, Thunnissen E, et al. Non-classic EGFR mutations in a cohort of Dutch EGFR-mutated NSCLC patients and outcomes following EGFR-TKI treatment. 2016;115(12):1504–1512. | ||
Lohinai Z, Hoda MA, Fabian K, et al. Distinct epidemiology and clinical consequence of classic versus rare EGFR mutations in lung adenocarcinoma. J Thorac Oncol. 2015;10(5):738–746. | ||
Chen Y, Ye L, Stanford RR, Zhang D, Zhang X, Wei W. Distinct epithelial growth factor receptor mutation profile in non-small cell lung cancer patients from the Xuanwei area of China. Mol Clin Oncol. 2016;4(5):749–755. | ||
Midha A, Dearden S, McCormack R. EGFR mutation incidence in non-small cell lung cancer of adenocarcinoma histology: a systematic review and global map by ethnicity (mutMapII). Am J Cancer Res. 2015;5(9):2892–2911. | ||
Hosgood HD III, Pao W, Rothman N, et al. Driver mutations among never smoking female lung cancer tissues in China identify unique EGFR and KRAS mutation pattern associated with household coal burning. Respir Med. 2013;107(11):1755–1762. | ||
Kristina SA, Endarti D, Prabandari YS, Ahsan A, Thavorncharoensap M. Burden of cancers related to smoking among the Indonesian population: premature mortality costs and years of potential life lost. Asian Pac J Cancer Prev. 2015;16(16):6903–6908. | ||
World Health Organization. Global Adult Tobacco Survey: Indonesia Report 2011. Geneva: WHO; 2012. | ||
Wang Q, Mou J, Yang X, et al. EGFR mutations in patients with lung adenocarcinoma in southwest China: are G719S/A and L861Q more likely detected in tumors derived from smokers? Lung Cancer (Auckl). 2013;4:27–33. | ||
Suzuki K. Differences in EGFR and KRAS mutation spectra in lung adenocarcinoma of never and heavy smokers. Oncol Lett. 2013; 6(5):1–6. | ||
Li H, Hu H, Wang R, et al. Primary concomitant EGFR T790M mutation predicted worse prognosis in non-small cell lung cancer patients. Onco Targets Ther. 2013;7:513–524. | ||
Chen L-Y, Molina-Vila MA, Ruan S-Y, et al. Coexistence of EGFR T790M mutation and common activating mutations in pretreatment non-small cell lung cancer: a systematic review and meta-analysis. Lung Cancer. 2016;94:46–53. | ||
Soh J, Okumura N, Lockwood WW, et al. Oncogene mutations, copy number gains and mutant allele specific imbalance (MASI) frequently occur together in tumor cells. PLoS One. 2009;4(10):e7464. | ||
Oakley GJ, Chiosea SI. Higher dosage of the epidermal growth factor receptor mutant allele in lung adenocarcinoma correlates with younger age, stage IV at presentation, and poorer survival. J Thorac Oncol. 2011;6(8):1407–1412. | ||
Malapelle U, Vatrano S, Russo S, et al. EGFR mutant allelic-specific imbalance assessment in routine samples of non-small cell lung cancer. J Clin Pathol. 2015;68(9):739–741. | ||
Fujita Y, Suda K, Kimura H, et al. Highly sensitive detection of EGFR T790M mutation using colony hybridization predicts favorable prognosis of patients with lung cancer harboring activating EGFR mutation. J Thorac Oncol. 2012;7(11):1640–1644. | ||
Maheswaran S, Sequist LV, Nagrath S, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med. 2008; 359(4):366–377. | ||
Ramalingam S, Yang JCH, Lee CK, et al. LBA1_PR: osimertinib as first-line treatment for EGFR mutation-positive advanced NSCLC: updated efficacy and safety results from two Phase I expansion cohorts. J Thorac Oncol. 2016;11(4):S152. | ||
Goodwin PM. First-line osimertinib effective in T790m-mutated EGFR lung cancer. Oncol Times. 2016;38(12):35. | ||
Oxnard GR, Miller VA, Robson ME, et al. Screening for germline EGFR T790M mutations through lung cancer genotyping. J Thorac Oncol. 2012;7(6):1049–1052. | ||
Gazdar A, Robinson L, Oliver D, et al. Hereditary lung cancer syndrome targets never smokers with germline EGFR gene T790M mutations. J Thorac Oncol. 2014;9(4):456–463. | ||
Kobayashi Y, Azuma K, Nagai H, et al. Characterization of EGFR T790M, L792F, and C797S mutations as mechanisms of acquired resistance to afatinib in lung cancer. Mol Cancer Ther. 2017;16(2):357–364. | ||
Chen K, Zhou F, Shen W, et al. Novel mutations on EGFR Leu792 potentially correlate to acquired resistance to osimertinib in advanced NSCLC. J Thorac Oncol. 2017;12(6):e65–e68. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.