Back to Archived Journals » Research and Reports in Biology » Volume 7
Ultraviolet modification of Chlamydomonas reinhardtii for carbon capture
Authors Gopal N, Sudhakar K
Received 18 October 2015
Accepted for publication 28 January 2016
Published 15 April 2016 Volume 2016:7 Pages 41—46
DOI https://doi.org/10.2147/RRB.S98536
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Zvi Kelman
Nikhil S Gopal,1 K Sudhakar2
1The Lawrenceville School, Lawrenceville, NJ, USA; 2Bioenergy Laboratory, Malauna Azad National Institute of Technology, Bhopal, India
Purpose: Carbon dioxide (CO2) levels have been rising rapidly. Algae are single-cell organisms with highly efficient CO2 uptake mechanisms. Algae yield two to ten times more biomass versus terrestrial plants and can grow nearly anywhere. Large scale CO2 sequestration is not yet sustainable due to high amounts of nitrogen (N) and phosphate (P) needed to grow algae in media.
Methods: Mutant strains of Chlamydomonas reinhardtii were created using ultraviolet light (2.2–3 K J/m2) and natural selection using media with 20%–80% lower N and P compared to standard Sueoka's high salt medium. Strains were selected based upon growth in media concentrations varying from 20% to 80% less N/P compared to control. Biomass was compared to wild-type control (CC-125) using direct counts, optical density dry weight, and mean doubling time.
Results: Mean doubling time was 20 and 25 hours in the low N and N/P strains, respectively (vs 66 hours in control). Using direct counts, growth rates of mutant strains of low N and N/P cultures were not statistically different from control (P=0.37 and 0.70), respectively.
Conclusion: Two new strains of algae, as well as wild-type control, were able to grow while using 20%–40% less N and P. Ultraviolet light-based modification of algae is an inexpensive and alternative option to genetic engineering techniques. This technique might make larger scale biosequestration possible.
Keywords: biosequestration, ultraviolet, carbon sequestration, carbon capture, algae
Introduction
Carbon dioxide (CO2) levels in Earth’s atmosphere have been rising steadily. Due to the greenhouse effect of trapped CO2 in the atmosphere, this increase is believed to be causing global warming.1 Many species of algae absorb nutrients such as CO2, nitrogen (N), and phosphate (P) as part of normal growth. While other plants can also absorb CO2, algae are two to ten times more efficient at absorbing CO2 and produce 15–300 times more biomass than land-based plants.2 Algae can be grown in many climates and can be harvested year-round instead of seasonally. It has been proposed that algae can be used to absorb CO2 near emission sources and buried as biomass to reduce our carbon footprint.3
However, simply cultivating algae in large open pools to sequester CO2 emissions would not be sustainable over the long term as valuable N and P would need to be added on a continual basis.4 Calculations estimate that large scale biosequestration using algae would require 88 million tons of rock P per year, where as annual US production is estimated to be only 40 million tons.5
A way to sequester CO2 without negatively impacting N and P levels is needed. Currently, no strains of algae exist that can sequester CO2 with minimal uptake of N and P. Even a small reduction in N or P requirements for algae might make carbon sequestration using algae on a large scale feasible.
Genetic engineering techniques have been used to create modified strains of algae.6 However, genetic modification is time intensive, requires expensive equipment, and raises questions about transfer of antibiotic-resistant genes. Ultraviolet (UV) light is mostly used to disinfect microorganisms, but it has also been used to create algae strains with interesting properties by damaging the DNA structure. The use of UV light to create new strains of algae deficient in both N/P uptake has never been attempted.
The primary objective of this experiment was to create new strains of Chlamydomonas reinhardtii using UV light that would grow and sequester CO2 as well as wild-type control strains while using less N and P.
Materials and methods
Algae growth
Growth media were prepared using Sueoka’s high salt media.7 A stock solution of N salts (based upon ammonium chloride) was added to stock solution of P salts (potassium phosphate dibasic and potassium phosphate monobasic). The concentrations of the N and P sources were adjusted as a percentage of the standard (noted as 100%). For example, an 80% N solution required 80% of 10.0 g of ammonium chloride (or 8.0 g). Eight different concentrations of the P media were used as shown in Table 1. A pH meter was used to check the final pH of growth media (target range 7.0) before adding algae strains. New culture flasks were created using growth media and an aliquot (10 mL) of the original culture before destruction or measurement of dry weight.
  | Table 1 Nitrogen (N) and phosphate (P) dilution factors used in growth media |
C. reinhardtii algae cultures were purchased from the Chlamydomonas Resource Center (University of Minnesota, St. Paul, MN, USA). The cultures were grown in flasks with a timer attached to a fluorescent lamp, simulating a 16-hour light/8-hour dark cycle. Ambient air was bubbled into each flask using an aquarium air pump and tubing. Once satisfactory growth in the exponential phase was achieved, UV light using a Phillips TUV T8 lamp was used to induce DNA damage using the method of Nichols.8 The following intensities of UV light were used: 2.2, 120, 700, or 3,000 J/m2. Using natural selection, the surviving strains were grown in various low N conditions (20%, 40%, 60%, or 80%) to allow any beneficial mutations to exhibit themselves. Once a strain was identified that grew well under these low N conditions, another round of UV light exposure and natural selection using varying concentrations of low N and P media was conducted (20%–80% P). This process was repeated until strains emerged with optimal growth characteristics under low nutrient conditions.
Biomass measurements
The biomass of the strains was measured using hemacytometer cell counts, gravimetric dry weight, and a spectrophotometer. A well-mixed sample of algae was collected, fixed with iodine tincture, and counted in triplicate on a hemacytometer using a 100× compound microscope. All the algae cells within a defined square of the hemacytometer were manually counted and multiplied by a dilution factor to arrive at a cell concentration. Another aliquot of this sample was used to measure optical density at wavelength 550 nm on a Spectronic 20 spectrophotometer using a 1.5 mL plastic cuvette. Gravimetric dry weight was calculated by measuring the volume of an algae culture and then passed through an 8 mm Whatman filter paper held in a glass funnel. The filter paper was weighed before and after drying in a low temperature oven overnight (60°C).
Generation time (g) was calculated based upon the following formula:9
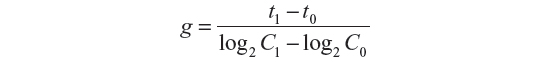
where t1, time1; t0, time 0; C1, cell density at t1; and C0, cell density at t0.
Control, sample size, and statistics
A control strain was included based upon recommendation from the curator at the Chlamydomonas Resource Center. A standard wild-type strain upon which most genetic studies are based was chosen as the control, CC-125.
The sample size of eight points per growth curve was based upon an online calculator using the following factors: 5% significance, 6-point standard deviation, 50% power, and a 12-hour difference in mean generation time.10
The growth of algae is an exponential function, and each time point is dependent upon the concentration of the prior reading. Typically, independence of values from one point to the next is required for most parametric statistical testing. Since the values of cell counts within one strain were not independent from each other, a nonparametric test had to be used. The analyses were based upon a statistical method to compare viral cultures having similar growth characteristics as algae.11 The null hypothesis was that there was no difference between the test and control strains as measured via the growth curve. If the P-value was ≥0.05, then the null hypothesis would be rejected and it can be concluded that there was no statistical difference between the curves. First, a Kruskal–Wallis test was used to look at overall differences in the growth curves. A Mann–Whitney U test was then used to compare individual growth curves against each other. Two-tailed P-values were calculated and considered significant if <0.05. Growth curves were plotted using Excel scatter plots and fitted with an exponential trendline. R2 values for each of the curves were also calculated using Excel. These statistical methods allowed for a comparison of the various growth strains against each other and, most importantly, against the control.
Results
Algae dry weights
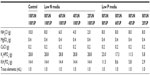
The mean dry weight in mg/mL for each of the algae strains is presented in Table 2. Variability was high and may be due to imprecision of the analytical balance used.
  | Table 2 Dry weight of algae strains |
Doubling time
Doubling time (or generation time) was calculated using time zero and each subsequent sampling time point (roughly once a day). The average doubling time for each strain is presented in Table 3. The most rapid growth was seen in the modified low N strain (60%N/100%P) and low N/P strain (80%N/80%P) with doubling times of 18 and 25 hours, respectively.
  | Table 3 Growth rates of strains |
Optical density
A scatter-plot of the relationship between optical density and cell count is provided in Figure 1. The readings were quite variable at the low and high ends of the transmittance readings. The most reliable readings were between 40% and 80% transmittance.
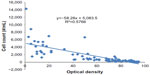  | Figure 1 Correlation between optical density and cell counts. |
Cell counts and growth curve comparisons
Mean values for cell counts (triplicate) at each time point are shown in Table 4. The values are presented as mean value in mg/mL after the appropriate dilution factor.
  | Table 4 Mean of triplicate cell counts (#/mL) over time |
The growth curves based upon the manual cell counts for the control and two best growing strains are provided in Figure 2. The curves show that all three strains grow in an exponential manner. The fitted exponential lines had excellent correlation (R2 of 0.8–0.9 range).
The results from the statistical tests are shown in Table 5. The Kruskal–Wallis test found no difference overall among all three curves (K=1.700, P=0.427). Similarly, the individual comparisons of the growth curves versus control were not statistically different using the Mann–Whitney U test (P=0.374 and 0.700). The null hypothesis was, therefore, rejected and it was concluded that there was no statistically significant difference among the new UV-irradiated strains and control.
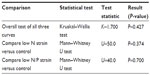  | Table 5 Statistical test results |
Description of mutant strain
A microscopic image of both mutant and control strains (CC-125) is provided under 100× magnification in Figure 3.
  | Figure 3 Microscopic image (100×) comparing wild-type and UV-modified strains. |
Discussion
Although many of the algae strains created using UV light grew poorly, two cultures grew quite well. The low N strain grew in the presence of low N (20%–40% less than required in control). Similarly, the low N/P strain grew in the presence of low N and low P (20% less N and P than control). These differences were not statistically different from the control wild-type strain CC-125.
These findings have several limitations. Direct cell counts were the most accurate measurement for biomass, and other methods (optical density and dry weight) were less reliable. The average cell size and/or cell volume differed between strains. The calculation of dry weight is important to quantify biomass but requires destruction of the algae culture. It would be important for future work to verify that these strains can sequester CO2 as well as control using other methods (radiolabelled carbon, ash-free dry weight, etc). The growth curves were replicated only in triplicate and for up to 250 hours. Longer growth and with more frequent measurement would be important. Measurement of N and P uptake would also be important for future work. Another limitation is the use of the Mann–Whitney U test for multiple comparisons; alternatives should be considered for future work.
The study done here shows possible improvement in photosynthetic productivity with mutant type UV light exposed cultures and possessing a lower level of light-harvesting pigment. Genetic engineering tools, coupled with sensitive absorbance, may, in principle, alleviate overabsorption of incident light by individual cells in high-density cultures. This in turn might help minimize dissipation of irradiance. This might also diminish the cell shading that occurs with normally pigmented wild-type cells, thus allowing a more uniform illumination of the whole cells, especially in cultures characterized by high biomass densities. Additionally, it is quite possible that these improvements can be extended to the efficiency of carbon fixation reactions and biofuel production.
These new strains of algae could be grown in large pools near sources of CO2 emission and be a self-sustaining CO2 sink. A cycle could be created in algae pools to continuously absorb CO2 until maximum capacity is reached, then the algae could be collected, dried, and buried underground.
It is estimated that algae could capture 95 million tons of CO2/ha/year.12 These newly created strains require 20%–40% less N and P to grow. Assuming that all the resources saved by needing 20%–40% less N and phosphorus are reinvested back into growing more algae, an extra 19–38 million tons of CO2/ha could be captured each year.
To put this into perspective, let us assume that ten large algae capture facilities are built (each at 10,000 acres absorbing 35 million tons of CO2 per year). Using these mutant strains of algae could allow an extra 14 million tons of CO2 to be absorbed each year. This is equivalent to the amount of CO2 that 289 million trees absorb over a 10-year period.13 Assuming a tree density of 300 trees/acre, this is an area slightly larger than the size of Rhode Island. Further modifications in algae CO2, and P and N pathways would be possible, potentially allowing for even greater efficiency in CO2 storing ability.
Conclusion
The study was carried out using mutant strains of C. reinhardtii to evaluate the growth rate, doubling time, and potential for carbon capture. The following conclusions were drawn from the study. These new strains of algae were created using a relatively simple method using UV light which could grow in media with 20%–40% less N and P compared to control. The current findings reiterate and support the potential of genetically modified microalgae for CO2 fixation. Further studies on the optimization of medium components and light growing conditions to enhance the growth yield of the culture have to be carried out. Extension of this method may enable further refinements in reducing N and P requirements to help make large scale biosequestration using algae feasible.
Acknowledgments
Funding was provided by Johns Hopkins Center for Talented Youth – Cogito Award. Thanks to Dr Elizabeth Fox and John Schiel at the Lawrenceville School for providing assistance and Matt Laudon at the Chlamydomonas Resource Center for providing advice on algae strains.
Author contributions
NSG devised the concepts, executed the research plan, and analyzed the data. KS reviewed the analyses and manuscript and provided overall guidance. All authors contributed toward data analysis, drafting and revising the paper and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
Solomon S, Plattner G-K, Knutti R, Friedlingstein P. Irreversible climate change due to carbon dioxide emissions. Proc Natl Acad Sci U S A. 2009;106(6):1704–1709. | |
Chisti Y. Biodiesel from microalgae beats bioethanol. Trends Biotechnol. 2008;26(3):126–131. | |
Zeiler KG, Heacox DA, Toon ST, Kadam KL, Brown LM. The use of microalgae for assimilation and utilization of carbon dioxide from fossil fuel-fired power plant flue gas. Energ Convers Manage. 1995;36(6):707–712. | |
Sayre R. Microalgae: the potential for carbon capture. Bioscience. 2010;60(9):722–727. | |
Rhodes C. Why Algal Biofuelds May Never Hold the Key to the Future. Oil and Energy Insider 2012; February 12, 2012. Available from: http://oilprice.com/Alternative-Energy/Biofuels/Why-Algal-Biofuels-May-Never-Hold-the-Key-to-the-Future.html. Accessed September 30, 2014. | |
Takahashi Y, Utsumi K, Yamamoto Y, Hatano A, Satoh K. Genetic engineering of the processing site of D1 precursor protein of photosystem II reaction center in Chlamydomonas reinhardtii. Plant Cell Physiol. 1996;37(2):161–168. | |
Sueoka N. Mitotic replication of deoxyribonucleic acid in Chlamydomonas reinhardi. Proc Natl Acad Sci U S A. 1960; 46(1):83–91. | |
Nichols G, Syrett P. Nitrate reductase deficient mutants of Chlamydomonas reinhardii. Isolation and genetics. J Gen Microbiol. 1978; 108(1):71–77. | |
Sager R, Granick S. Nutritional studies with Chlamydomonas reinhardi. Ann N Y Acad Sci. 1953;56(5):831–838. | |
Schoenfeld DA. Statistical considerations for clinical trials and scientific experiments; 2010. Available from: http://hedwig.mgh.harvard.edu/sample_size/size.html. Accessed September 30, 2014. | |
Wang GP, Bushman FD. A statistical method for comparing viral growth curves. J Virol Methods. 2006; 135(1):118–123. | |
Sudhakar K, Premalatha M. Theoretical assessment of algal biomass potential for carbon mitigation and biofuel production. IJEE. 2012;3(3):232–240. | |
Environmental Protection Agency Climate Change Calculator. Retrieved on February 8, 2015. Available from: http://www.epa.gov/cleanenergy/energy-resources/calculator.html. Accessed on February 8th, 2015. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

