Back to Journals » Cancer Management and Research » Volume 11
UBE2T promotes proliferation via G2/M checkpoint in hepatocellular carcinoma
Authors Liu LL, Zhu JM , Yu XN, Zhu HR, Shi X, Bilegsaikhan E, Guo HY, Wu J, Shen XZ
Received 23 January 2019
Accepted for publication 19 August 2019
Published 13 September 2019 Volume 2019:11 Pages 8359—8370
DOI https://doi.org/10.2147/CMAR.S202631
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Beicheng Sun
Li-Li Liu,1,2,* Ji-Min Zhu,1,2,* Xiang-Nan Yu,1,2 Hai-Rong Zhu,1,2 Xuan Shi,1,2 Enkhnaran Bilegsaikhan,1,2 Hong-Ying Guo,1,2 Jian Wu,1–3 Xi-Zhong Shen1–3
1Department of Gastroenterology and Hepatology, Zhongshan Hospital, Fudan University, Shanghai 200032, People’s Republic of China; 2Shanghai Institute of Liver Diseases, Zhongshan Hospital, Shanghai Medical College, Fudan University, Shanghai 200032, People’s Republic of China; 3Department of Medical Microbiology and Parasitology, Ministry of Education/National Health Commission/Chinese Academy of Medical Sciences, Key Laboratory of Medical Molecular Virology, School of Basic Medical Sciences, Shanghai Medical College, Fudan University, Shanghai 200032, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Xi-Zhong Shen
Department of Gastroenterology and Hepatology, Zhongshan Hospital of Fudan University, 180 Fenglin Road, Shanghai 200032, People’s Republic of China
Tel +86 216 404 1990
Email [email protected]
Jian Wu
Department of Medical Microbiology and Parasitology, School of Basic Medical Sciences, Shanghai Medical College, Fudan University, 138 Yixue Yuan Road, P.O. Box 228, Shanghai 200032, People’s Republic of China
Tel +86 21 5423 7705
Email [email protected]
Background: Growing evidence suggests that the ubiquitin-proteasome system is involved in the pathogenesis and recurrence of hepatocellular carcinoma (HCC); yet, little is known about the role of ubiquitin-conjugating enzyme E2T (UBE2T) in HCC.
Materials and methods: UBE2T levels were detected in HCC tissues and hepatoma cell lines using quantitative reserve transcriptase-polymerase chain reaction and Western blot analysis. Next, the changes of phenotypes after UBE2T knockdown or overexpression were evaluated using in vitro methods. Finally, the mechanism of UBE2T in HCC was tested using ex vivo and in vivo methods.
Results: In the present study, we reported that UBE2T mRNA and protein levels were significantly upregulated in HCC tissues compared to adjacent non-tumor tissues. Additionally, suppression of UBE2T expression inhibited proliferation, colony formation, tumorigenesis, migration, and invasion of hepatoma cells, whereas UBE2T overexpression led to the opposite outcomes. Moreover, suppression of UBE2T expression resulted in an increase in G2/M phase and a decrease in the percentage of cells in G1 phase, which indicated a cell cycle arrest at the G2/M phase. In contrast, the percentage of cells in G2/M phase decreased following UBE2T overexpression. Further study indicated that UBE2T regulated the G2/M transition by modulating cyclin B1 and cyclin-dependent kinase 1.
Conclusion: Taken together, the findings of the present study uncover biological functions of UBE2T in hepatoma cells, and delineate preliminary molecular mechanisms of UBE2T in modulating HCC development and progression.
Keywords: hepatocellular carcinoma, UBE2T, ubiquitination, cell cycle
Introduction
Hepatocellular carcinoma (HCC) is the most common form of liver cancer, and it is one of the most prevalent malignancies that threatens human health.1,2 The disease develops most often from exposure to risk factors, including viral hepatitis infection, obesity, chronic ethanol consumption, and aflatoxin ingestion.3 The molecular mechanism underlying HCC development remains poorly understood, although several oncogenes and tumor suppressors have been implicated in the initiation and development of HCC.
Growing evidence suggests that the ubiquitin-proteasome system (UPS) is involved in HCC pathogenesis and recurrence.4 The UPS is a degradation pathway for most proteins in eukaryotic cells. It participates in various biological processes and signaling pathways, including cell cycle, DNA repair, transcription regulation, apoptosis, and immune response.5,6 The UPS contains six components: proteasome, ubiquitin (Ub), ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2), ubiquitin ligase (E3), and deubiquitin enzyme.7 E2, a carrier for ubiquitin (Ub) delivery, is an essential component in the UPS. It plays a crucial role in Ub attachment to cellular proteins.8 Several E2s, such as ubiquitin-conjugating enzyme 2C (UBE2C), have been shown to be involved in cell cycle modulation, thus playing an essential role in proliferation of tumors. UBE2C cooperates with E3 ligase anaphase-promoting complex/cyclosome to regulate cell cycle progression.9,10 Ubiquitin-conjugating enzyme E2T (UBE2T), also known as HSPC150 or FANCT, was first identified as a key factor in the Fanconi anemia pathway.11 It is an E2 enzyme that plays a role in cell proliferation, and it is required for mono-ubiquitination of FANCD2 and the maintenance of chromosome stability. Several studies have identified UBE2T as an oncogene in multiple types of malignancies, including lung, breast, bladder, gastric, prostate cancer, and HCC.12–17 However, the role of UBE2T in modulating HCC initiation, progression, and metastasis is still unclear.
In this study, our goal was to determine the mechanisms by which UBE2T affects HCC tumorigenesis. Our findings demonstrate that UBE2T exerts promoting effects on proliferation, colony formation, migration, and invasion of hepatoma cells through regulating G2/M transition, and these findings underscore the critical role of UBE2T in hepatocarcinogenesis.
Materials and methods
Cell culture
Huh-7 cell line was kindly provided by Professor Mark Feitelson, PhD from Temple University, Philadelphia, PA, USA in 2005. SK-Hep1, HepG2 cells were obtained from the American Type Culture Collection (Manassas, VA, USA) in 2003. SMCC-7721, MHCC-97H, and BEL-7402 were obtained from the Institute of Liver Cancer, Zhongshan Hospital of Fudan University (Shanghai, People’s Republic of China). Immortalized human fetal hepatocyte (FH) cell linewas generated in Dr Mark Zern’s lab.18,19 The use of human cell lines and liver tissues was approved by the Institutional Review Board of Zhongshan Hospital, Fudan University. All cell lines used in the present study were authenticated for accuracy by short tandem repeat (STR) test in December 2018 by GENEWIZ, Inc., Suzhou, People’s Republic of China, and tested periodically for mycoplasma by PCR. Four HCC cell lines (including SMCC-7721, Huh-7, BEL-7402, and MHCC-97H) and human embryonic kidney cell line HEK293T were cultured in Dulbecco’s modified Eagle medium (DMEM; Hyclone, Logan, UT, USA) supplemented with 10% (v/v) fetal bovine serum (FBS; Gibco, Gaithersburg, MD, USA) and 1% (v/v) penicillin/streptomycin. Two HCC cell lines (including SK-Hep1 and HepG2) and FH were cultured in minimal Eagle medium (Gibco) supplemented with 10% (v/v) FBS, 1% (v/v) penicillin/streptomycin, 1% (v/v) L-glutamine, and 1% (v/v) sodium pyruvate (all from Life Technologies, Grand Island, NY, USA). All cell lines were cultured in a humidified incubator at 37°C with 5% CO2.
Quantitative reserve transcriptase-polymerase chain reaction (qRT-PCR)
RNA was extracted from tumor specimens using Trizol reagent (Invitrogen Corp., Carlsbad, CA, USA). cDNA was synthesized using PrimeScript™ RT reagent Kit (Takara, Kusatsu, Japan). qRT-PCR was performed with the SYBR® Premix TM kit (Invitrogen). Conditions for RT-PCR were as follows: 95°C for 10 mins, 40 cycles of 94°C for 30 s, 60°C for 30 s, 72°C for 30 s, and a final extension of 10 mins at 72°C. The primers for amplification were as follows: UBE2T-Forward: 5ʹ-ATCCC TCAAC ATCGC AACTG T-3ʹ, UBE2T-reverse: 5ʹ-CAGCC TCTGG TAGAT TATCA AGC-3ʹ, glyceraldehyde 3-phosphate dehydrogenase (GAPDH)-Forward: 5ʹ-GAAGA TGGTG ATGGG ATTTC-3ʹ, GAPDH-reverse: 5ʹ-GAAGG TGAAG GTCGG AGTC-3ʹ. All experiments were performed independently for three times.
Western blot analysis
Cells were lysed using RIPA lysis buffer (Beyotime, Shanghai, People’s Republic of China). Protein samples were separated using 4–20% gradient SDS-PAGE, and then transferred onto a polyvinylidene fluoride membrane. The membrane was incubated with a primary antibody at 4°C overnight followed by a horseradish peroxidase-conjugated secondary antibody for 2 hrs at room temperature. Enhanced chemiluminescence (ECL) reagents were used to visualize the bands. The following primary antibodies were used: anti-UBE2T (Proteintech, Chicago, IL, USA), anti-GAPDH (Proteintech), anti-cyclin B1 (Cell Signaling Technology, Danvers, MA, USA), anti-cyclin-dependent kinase 1 (CDK1) (Cell Signaling Technology), anti-p21 (Proteintech), and anti-p27 (Proteintech).
Establishment of UBE2T knockdown and overexpression in HCC cell lines
The HCC cell lines were stably transfected with UBE2T shRNA or overexpression vector using Lipofectamine 3000 (Invitrogen). Briefly, HCC cells were seeded in 6-well plates the day before transfection. After 24 hrs, the four types of lentiviruses were separately added to HCC cells with normal culture medium and 5 μg/mL Polybrene (Sigma-Aldrich, St. Louis, MO, USA). Twelve hours later, the medium was removed and replaced with fresh culture medium containing 10% FBS and puromycin. One week later, transduced cells were collected for subsequent culture. UBE2T expression was validated using Western blot analysis. The shRNA sequence targeting the human UBE2T is 5ʹ-TGAGG AAGAG ATGCT TGATA A-3ʹ. The shRNA against UBE2T and the UBE2T cDNA was synthesized by Sango Biotech (Shanghai, People’s Republic of China). The following names were used to define stably transfected cell types: UBE2T knockdown (UBE2T-KD), UBE2T overexpression (UBE2T-OE), control cells transfected with the control vector (KD-control), and control cells transfected the empty vector (OE-control).
Cell proliferation assay
Cell proliferation was evaluated using a CCK8 Kit (Beyotime) according to the manufacturer’s instruction. Stably transfected HCC cells and control cells were seeded in 96-well plates in triplicate at an initial density of 2.5×103 cells per well. After being cultured for 24, 48, 72, and 96 hrs, CCK-8 solution in 10 μL was added to each well, and cells were further incubated at 37°C for 2 hrs. Optical density (OD) value was measured spectrophotometrically at 450 nm wavelength.
Assays of colony formation and soft agar colony formation
Stably transfected cells were plated in 6-well plates (1×103 cells per well), and then cultured for 14 days. For an assay of soft agar colony formation, 1×103 cells were suspended in 1 mL of complete medium containing 0.3% agarose (Sigma-Aldrich). The agarose-cell mixture was placed onto the top layer of solidified 0.6% complete medium/agar. After 14 days of culture, colonies were fixed with 4% paraformaldehyde for 20 mins, stained with 0.1% crystal violet for 15 mins, and then washed with phosphate-buffered saline (PBS) before counting. Viable colonies that were larger than 0.5 mm were counted.
Assays of transwell migration and invasion
A 24-well plate containing 8-mm pore size chamber inserts (Corning, NY, USA) was used to evaluate the migration and invasion of HCC cells. Cells were harvested and resuspended in FBS-free medium. For the migration assay, 2×104 cells were seeded in the upper chamber. For the invasion assay, the membrane was coated with Matrigel (BD Pharmingen, San Diego, CA, USA) to form a matrix barrier, and 4×104 cells were seeded in the upper chamber. Five hundred microliter of DMEM with 10% FBS was added in each lower chamber. After 24 and 48 hrs of incubation at 37°C, cells migrated through the membrane pore were fixed by 4% paraformaldehyde and stained with 0.1% crystal violet. Finally, stained cells were photographed and counted using a microscope.
Flow cytometry analysis for cell cycle and apoptosis
Cells from a 6-well plate were harvested and fixed in 70% cold ethanol in PBS after washing in cold PBS. After incubation with 70% ethanol at 4°C overnight, cells were pelleted in a cooled centrifuge and washed with cold PBS again. Then, 500 μL propidium iodide (BD Pharmingen) was added to 1×105 cells for 20 mins at room temperature. A total of 2×104 cells were analyzed using a BD FACS Canto for cell cycle and the data were analyzed using ModFit LT Software.
To detect apoptosis, an Annexin V Apoptosis Detection Kit (BD Pharmingen) was used according to the manufacturer’s instructions. The Annexin V-FITC and PI fluorescence levels were measured by flow cytometry (FACS Calibur, BD Biosciences, USA). Annexin V-positive cells (both PI-negative and -positive) were defined as apoptotic cells.
Xenograft tumor model
BALB/c nude mice (4–5 weeks old, 15–16 g) were purchased from SLAC Laboratory Animal Co. Ltd (Shanghai, People’s Republic of China). The mice were housed in wire-bottomed cages with a 12-hrs light-dark cycle, 21–23°C ambient temperature, and were offered food and water ad libitum. SMCC-7721 cells at 8×106 (UBE2T knockdown and control) in 100 μL of PBS were injected separately into flanks of nude mice subcutaneously. Length (L) and width (W) of tumors were measured with calipers every 3 days, and tumor volume (V) was calculated using the following equation: V=L×W2/2. On day 21, tumor-bearing mice were anesthetized, and tumors were excised, weighed, serially sliced, and stained with anti-UBE2T antibody and anti-Ki67 antibody. Animal care and procedures were approved by the Institutional Animal Care and Use Committee of Zhongshan Hospital, Fudan University, and all procedures were conducted following the NIH Guidelines of Experimental Animal Handling and Use.
Immunohistochemistry (IHC)
Formalin-fixed paraffin-embedded tissues were cut into 4-μm sections and rehydrated in a graded series of alcohol. Endogenous peroxidase activity was blocked with 3% H2O2. Subsequently, sections were steamed for 3 mins for antigen retrieval in 10 mM citrate buffer (pH 6.0) and incubated with a primary antibody at 4°C overnight. Then, tissue sections were incubated with a secondary antibody (Dako) at room temperature for 1 hr. The sections were stained with 3,3ʹ-diaminobenzidine tetrahydrochloride (DAB; Dako, Glostrup, Denmark) to visualize the stained areas and counterstained with hematoxylin.
Statistical analysis
All experimental operations were independently repeated for at least three times. Densitometry of the immunoblotting images was analyzed using Image J Software 1.51 (National Institutes of Health, Bethesda, MD, USA). The graphs were plotted using GraphPad Prism Software 7 (GraphPad Software, La Jolla, CA, USA). Statistical analysis was performed using SPSS 22.0 (SPSS, Inc., Chicago, IL, USA). Continuous data were shown as the mean±SD. For data that had a normal distribution and homogeneity of variance, two-tailed Student’s t test was performed to evaluate significant differences between two groups. P-values <0.05 were considered statistically significant.
Results
UBE2T is upregulated in HCC tissues and hepatoma cell lines
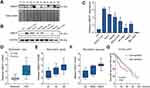
In our previous microarray study, we found that UBE2T was upregulated in HCC tissues compared with adjacent non-tumor tissues (unpublished data). In this study, we demonstrated that UBE2T is highly expressed in 5 of 6 HCC tissues (T) compared with adjacent non-tumor tissues using Western blot analysis (Figure 1A). We further confirmed the upregulation of UBE2T protein expression in seven hepatoma cell lines using immortalized human FHs as a control. The results showed that UBE2T protein expression was upregulated up to 1.3- to 6.4-fold in six hepatoma cell lines compared to FH (Figure 1B and C). Upregulation of UBE2T was also confirmed in the Wurmbach liver study in the oncomine database (Figure 1D). The expression level of UBE2T was correlated with the pathological grade and vascular invasion of human HCC tissue (Figure 1E and F). The survival data from the The Cancer Genome Atlas (TCGA) cohort showed that high expression of UBE2T was negatively correlated to a poor survival in HCC patients (Figure 1G). Taken together, our results suggested that UBE2T may act as an oncogene in HCC.
UBE2T promotes proliferation of hepatoma cells
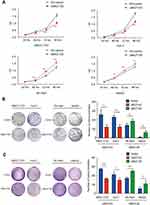
To further examine the role of UBE2T in HCC, we inhibited its expression in SMCC-7721 and Huh-7 cell lines by RNA inference (RNAi) approach and upregulated it in SK-Hep1 and HepG2 cell lines using a lentiviral transduction with the full sequence of UBE2T cDNA. We design three interference fragments of shUBE2T (sh1, sh2, and sh3). Western blot tests showed that the fragment of sh1 had the highest interference efficiency. Thus, we selected sh1 in the following experiments (Figure S1). The results of qRT-PCR and Western blot assays confirmed that transduction of lentiviral vector containing shRNA against UBE2T dramatically decreased UBE2T gene expression in SMCC-7721 and Huh-7 cells (approximately 80% decrease, P<0.05); whereas UBE2T overexpression by lentiviral transduction significantly increased UBE2T expression in SK-Hep1 and HepG2 cells (10- to 20-fold increase, P<0.05; Figure S2).
A CCK8 assay was conducted to examine the effect of UBE2T expression on cell proliferation. UBE2T knockdown significantly decreased the SMCC-7721 and Huh-7 cell numbers, while UBE2T overexpression increased the SK-Hep1 and HepG2 cell numbers (Figure 2A). To further confirm the role of UBE2T in tumorigenesis, we performed colony formation assay and soft agar colony formation assay with stable UBE2T-KD cells and corresponding controls, as well as UBE2T-OE and corresponding controls. The colony formation of hepatoma cells with UBE2T-KD was much lower than that of controls, while UBE2T overexpression markedly enhanced colony formation compared to controls (Figure 2B and C). In summary, these data demonstrate that UBE2T may exert a promoting role in cell proliferation and tumor formation.
UBE2T accelerates migration and invasion of hepatoma cells
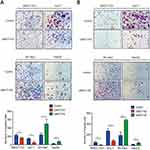
We performed a Transwell assay to confirm the role of UBE2T in migration and invasion in HCC. Downregulation of UBE2T significantly inhibited the migration ability of hepatoma cells, while overexpression of UBE2T significantly enhanced migration and invasion ability of hepatoma cells (Figure 3A, P<0.05). Furthermore, cell invasion was strongly inhibited in UBE2T-KD compared with controls, while UBE2T-OE possessed an increased cell invasion rate compared with controls (P<0.01; Figure 3B). These data suggest that UBE2T promotes HCC cell migration and invasion.
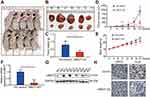
UBE2T regulates the tumorigenesis of HCC in nude mice
To confirm whether the growth-promoting effects of UBE2T are relevant to HCC growth in vivo, UBE2T-KD–treated SMCC-7721 cells and control cells were subcutaneously injected into BALB/c nude mice (Figure 4A and B). Knockdown of UBE2T resulted in an approximately 60% reduction in weight and 70% reduction in volume of xenograft tumors as compared to controls (Figure 4C and D; P<0.05). The nude mice growth curves are shown in Figure 4E. qRT-PCR and Western blot assay confirmed that UBE2T mRNA and protein levels were significantly lower in UBE2T-KD than in controls (Figure 4F and G). Also, mRNA and protein levels of UBE2T and Ki67 were decreased in xenograft tumors formed by UBE2T-KD cells by IHC (Figure 4H). Collectively, these results indicate that UBE2T may promote hepatoma cell proliferation in vivo.
UBE2T is involved in the transition from G2 to M by modulating cyclin B1 and CDK1 in cell cycle
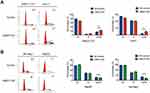
An alteration in proliferation is usually caused by change in cell cycle or apoptosis. Flow cytometry was performed to examine the effects of UBE2T on cell cycle progression and apoptosis. Cell cycle tests showed that suppressing UBE2T expression resulted in a significant increase in the percentage of cells in G2/M phase and a decrease in the percentage of cells in G1 phase, which indicated that downregulation of UBE2T promotes cell cycle arrest at G2/M phase (Figure 5A). In contrast, the cell portion in G2/M phase was decreased after upregulation of UBE2T (Figure 5B). However, UBE2T knockdown or overexpression had no significant effect on apoptosis (Figure S3). Taken together, these data indicate that UBE2T may facilitate a G2-to-M transition in hepatoma cells.
To investigate the potential mechanism of the proliferation-promoting function of UBE2T, cell cycle-related proteins were analyzed using Western blotting. The results revealed that cell cycle promoting factors, including cyclin B1 and CDK1, were upregulated in UBE2T-transfected cells. However, the cell cycle inhibitors p21Cip1 and p27Kip1 were also increased in UBE2T-OE cells but decreased in UBE2T-KD cells (Figure 6A and B). We also analyzed gene expression correlations between UBE2T and CCNB1 or CDK1 using a bioinformatic tool GEPIA (http://gepia.cancer-pku.cn/). The results showed that UBE2T mRNA was significantly correlated positively with mRNA levels of CCNB1 and CDK1 (Spearman coefficient R=0.88 and 0.92, respectively; P<0.05, Figure 6C and D). These findings suggest that UBE2T regulates G2/M transition by modulating cyclin B1 and CDK1 expression, and further controlling cell cycle and proliferation in hepatoma cells.
Discussion
UBE2T is a member of the E2 family that is required for the repair of damaged DNA.11,20 It is located at 1q32.1 and the gain of 1q is frequently observed in a variety of cancers.21 Recently, UBE2T upregulation has been demonstrated in different cancer, such as lung, breast, prostate, and gastric carcinoma. Several cancer cell lines exhibit UBE2T upregulation according to data mining in large-scale genome analysis and expression data in cBiopartal.22 In our present study, we demonstrated that UBE2T was remarkably upregulated in HCC tissues and hepatoma cell lines, which was consistent with previous findings, which found that UBE2T expression was negatively correlated with poor survival of HCC patients.13 Survival data from TCGA also supported this conclusion. Additionally, the prognostic value of UBE2T has been reported in breast, gastric, and nasopharyngeal carcinoma.16,23,24
Subsequent cell function test results showed that inhibition of UBE2T significantly represses proliferation, migration, and invasion of HCC cell lines as compared with controls, while UBE2T overexpression significantly promotes proliferation, migration, and invasion ability of HCC cells. Also, suppressing UBE2T expression by an RNAi approach inhibited tumor growth in a mouse model of HCC xenograft, which is consistent with previous studies.15,23,25 Overexpression of UBE2T exerted a promoting role in cell proliferation and induced epithelial-mesenchymal transition in gastric cancer cells.12 It also enhanced tumor growth in mouse models of prostate and nasopharyngeal cancer.15,23 Silencing of UBE2T inhibited proliferation and promoted apoptosis and cell cycle arrest in bladder cancer cells.14 In summary, it appears that UBE2T may exert a promoting effect on proliferation, invasion, and metastasis in various tumors.
Both cell cycle and apoptosis play a reciprocal role in modulating proliferation of tumor cells. Our results showed that G2/M phase arrest occurs after the suppression of UBE2T expression by an RNAi approach, in turn inhibiting the proliferation of hepatoma cells. These results were consistent with reports performed with bladder and gastric cancer cases.12,14 It was reported that interference with UBE2T downregulates cyclin D1 in gastric carcinoma cells, but did not provide any evidence to explain the G2/M arrest. The latest research by Liu et al found that UBE2T degrades p53 by ubiquitination and promotes proliferation in liver cancer cells.13 It is well known that p53 is an essential transcription factor for cell cycle regulation, and activation of the p53-DREAM pathway leads to cell cycle arrest. However, multiple signaling pathways are involved in downstream of p53, which may participate in the regulation of G1/S and G2/M checkpoints.26 Cyclin B1-CDK1 complex plays a central role in G2/M transition. Downregulation of CDK1 protein or inhibition of CDK1 kinase activity usually accompanied with the arrest of G2/M transition.27 Cyclin B1 and CDK1 protein levels were downregulated in the UBE2T-KD group, which is consistent with the arrest of G2/M transition. Our study is the first to find that UBE2T regulates G2/M transition through cyclin B1 and CDK1; however, the exact molecular interplay between UBE2T and the cyclin B1-CCK1 axis in during cell cycle control remains to be further investigated.
Simultaneously, we found that p21 and p27 were downregulated after knockdown of UBE2T, and vice versa. This result is inconsistent with previous studies.13 p21 and p27 belong to the Cip/Kip family of cyclin-dependent kinase inhibitors. It is generally believed that p21 and p27 act as tumor suppressors during tumorigenesis.28 However, growing evidence implies that p21 and p27 are oncogenic in various tumors with the mechanism of a reversal translocation from nucleus to cytosol.29–31 Our findings from UBE2T overexpressed hepatoma cells suggest that UBE2T may facilitate the translocation of p21 and p27 into the cytosol compartment.
UBE2T and its interaction with the E3 ligase FANCL in the Fanconi anemia have been well studied in recent years. UBE2T has a direct interaction with the BRCA1/BRAD1 complex in breast cancer. However, our study is unable to elucidate the direct protein–protein interaction of UBE2T with these complex in hepatoma cells. In the present study, we elucidated an indirect regulatory mechanism of UBE2T expression and molecular interaction. The E3 ligase that interacts with UBE2T has not been clarified yet in HCC, and further study remains to be needed. Tumor cells are more susceptible to proteasome inhibition due to their rapid division and disordered regulatory pathways. Proteasome inhibitors form a novel class of chemotherapeutic agents that lead to cell cycle arrest and cell death, though no specific UBE2T inhibitors are currently available for clinical use.
In conclusion, we carried out a relatively comprehensive study on UBE2T in HCC development and progression in vitro and in vivo. The findings demonstrate that UBE2T exerts promoting effects on proliferation, colony formation, migration, and invasion of hepatoma cells through regulation of G2/M transition in cell cycle. This study provides valuable insights into the underlying mechanisms of UBE2T in modulating the cyclin B1-CDK1 pathway, and points UBE2T as a promising target for HCC treatment.
Abbreviations
HCC, hepatocellular carcinoma; UPS, ubiquitin-proteasome system; Ub, ubiquitin; E1, the ubiquitin-activating enzyme; E2, the ubiquitin-conjugating enzyme; E3, the ubiquitin ligase; qPCR, quantitative real-time PCR; RNAi, RNA interference; UBE2T, ubiquitin-conjugating enzyme E2T; FDR, false discovery rate; UBD, ubiquitin-binding domain.
Acknowledgments
This study was supported by National Nature Science Foundation of China (grant numbers 1301820, 81402273, 81472673, 81572356, 81672720, 81672334, and 81871997) and Shanghai Science and Technology Commission (grant numbers 16ZR1406100 and 16140903700), as well as the Ministry of Science & Technology of China (#2016YFE0107400). Thanks to Dr Anna Williams for editing the manuscript.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi:10.3322/caac.20107
2. Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi:10.3322/caac.21262
3. Sherman M. Hepatocellular carcinoma: epidemiology, surveillance, and diagnosis. Semin Liver Dis. 2010;30(1):3–16. doi:10.1055/s-0030-1247128
4. Chen YJ, Wu H, Shen XZ. The ubiquitin-proteasome system and its potential application in hepatocellular carcinoma therapy. Cancer Lett. 2016;379(2):245–252. doi:10.1016/j.canlet.2015.06.023
5. Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet. 1996;30:405–439. doi:10.1146/annurev.genet.30.1.405
6. Hershko A, Ciechanover A. The ubiquitin system for protein degradation. Annu Rev Biochem. 1992;61:761–807. doi:10.1146/annurev.bi.61.070192.003553
7. Clague MJ, Heride C, Urbe S. The demographics of the ubiquitin system. Trends Cell Biol. 2015;25(7):417–426. doi:10.1016/j.tcb.2015.03.002
8. Stewart MD, Ritterhoff T, Klevit RE, Brzovic PS. E2 enzymes: more than just middle men. Cell Res. 2016;26(4):423–440. doi:10.1038/cr.2016.35
9. Hao Z, Zhang H, Cowell J. Ubiquitin-conjugating enzyme UBE2C: molecular biology, role in tumorigenesis, and potential as a biomarker. Tumour Biol. 2012;33(3):723–730. doi:10.1007/s13277-011-0291-1
10. Voutsadakis IA. Ubiquitin- and ubiquitin-like proteins-conjugating enzymes (E2s) in breast cancer. Mol Biol Rep. 2013;40(2):2019–2034. doi:10.1007/s11033-012-2261-0
11. Machida YJ, Machida Y, Chen Y, et al. UBE2T is the E2 in the Fanconi anemia pathway and undergoes negative autoregulation. Mol Cell. 2006;23(4):589–596. doi:10.1016/j.molcel.2006.06.024
12. Luo C, Yao Y, Yu Z, et al. UBE2T knockdown inhibits gastric cancer progression. Oncotarget. 2017;8(20):32639–32654. doi:10.18632/oncotarget.15947
13. Liu LP, Yang M, Peng QZ, et al. UBE2T promotes hepatocellular carcinoma cell growth via ubiquitination of p53. Biochem Biophys Res Commun. 2017;493(1):20–27. doi:10.1016/j.bbrc.2017.09.091
14. Gong YQ, Peng D, Ning XH, et al. UBE2T silencing suppresses proliferation and induces cell cycle arrest and apoptosis in bladder cancer cells. Oncol Lett. 2016;12(6):4485–4492. doi:10.3892/ol.2016.5237
15. Wen M, Kwon Y, Wang Y, Mao JH, Wei G. Elevated expression of UBE2T exhibits oncogenic properties in human prostate cancer. Oncotarget. 2015;6(28):25226–25239. doi:10.18632/oncotarget.4712
16. Ueki T, Park JH, Nishidate T, et al. Ubiquitination and downregulation of BRCA1 by ubiquitin-conjugating enzyme E2T overexpression in human breast cancer cells. Cancer Res. 2009;69(22):8752–8760. doi:10.1158/0008-5472.CAN-09-1809
17. Hao J, Xu A, Xie X, et al. Elevated expression of UBE2T in lung cancer tumors and cell lines. Tumour Biol. 2008;29(3):195–203. doi:10.1159/000148187
18. Wege H, Le HT, Chui MS, et al. Telomerase reconstitution immortalizes human fetal hepatocytes without disrupting their differentiation potential. Gastroenterology. 2003;124(2):432–444. doi:10.1053/gast.2003.50064
19. Zhou P, Lessa N, Estrada DC, et al. Decellularized liver matrix as a carrier for the transplantation of human fetal and primary hepatocytes in mice. Liver Transpl. 2011;17(4):418–427. doi:10.1002/lt.22270
20. Alpi A, Langevin F, Mosedale G, et al. UBE2T, the Fanconi anemia core complex, and FANCD2 are recruited independently to chromatin: a basis for the regulation of FANCD2 monoubiquitination. Mol Cell Biol. 2007;27(24):8421–8430. doi:10.1128/MCB.00504-07
21. Corson TW, Huang A, Tsao MS, Gallie BL. KIF14 is a candidate oncogene in the 1q minimal region of genomic gain in multiple cancers. Oncogene. 2005;24(30):4741–4753. doi:10.1038/sj.onc.1208641
22. Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science Signaling. 2013;6(269):pl1. doi:10.1126/scisignal.2004088
23. Hu W, Xiao L, Cao C, Hua S, Wu D. UBE2T promotes nasopharyngeal carcinoma cell proliferation, invasion, and metastasis by activating the AKT/GSK3beta/beta-catenin pathway. Oncotarget. 2016;7(12):15161–15172. doi:10.18632/oncotarget.7805
24. Yu H, Xiang P, Pan Q, et al. Ubiquitin-conjugating enzyme E2T is an independent prognostic factor and promotes gastric cancer progression. Tumour Biol. 2016;37(9):11723–11732. doi:10.1007/s13277-016-5020-3
25. Wang Y, Leng H, Chen H, et al. Knockdown of UBE2T inhibits osteosarcoma cell proliferation, migration, and invasion by suppressing the PI3K/Akt signaling pathway. Oncol Res. 2016;24(5):361–369. doi:10.3727/096504016X14685034103310
26. Engeland K. Cell cycle arrest through indirect transcriptional repression by p53: I have a DREAM. Cell Death Differ. 2018;25(1):114–132. doi:10.1038/cdd.2017.172
27. Sisinni L, Maddalena F, Condelli V, et al. TRAP1 controls cell cycle G2-M transition through the regulation of CDK1 and MAD2 expression/ubiquitination. J Pathol. 2017;243(1):123–134. doi:10.1002/path.4936
28. Jackson RJ, Adnane J, Coppola D, et al. Loss of the cell cycle inhibitors p21(Cip1) and p27(Kip1) enhances tumorigenesis in knockout mouse models. Oncogene. 2002;21(55):8486–8497. doi:10.1038/sj.onc.1205946
29. Ohkoshi S, Yano M, Matsuda Y. Oncogenic role of p21 in hepatocarcinogenesis suggests a new treatment strategy. World J Gastroenterol. 2015;21(42):12150–12156. doi:10.3748/wjg.v21.i42.12150
30. Warfel NA, El-Deiry WS. p21WAF1 and tumourigenesis: 20 years after. Curr Opin Oncol. 2013;25(1):52–58. doi:10.1097/CCO.0b013e32835b639e
31. Serres MP, Zlotek-Zlotkiewicz E, Concha C, et al. Cytoplasmic p27 is oncogenic and cooperates with Ras both in vivo and in vitro. Oncogene. 2011;30(25):2846–2858. doi:10.1038/onc.2011.9
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.